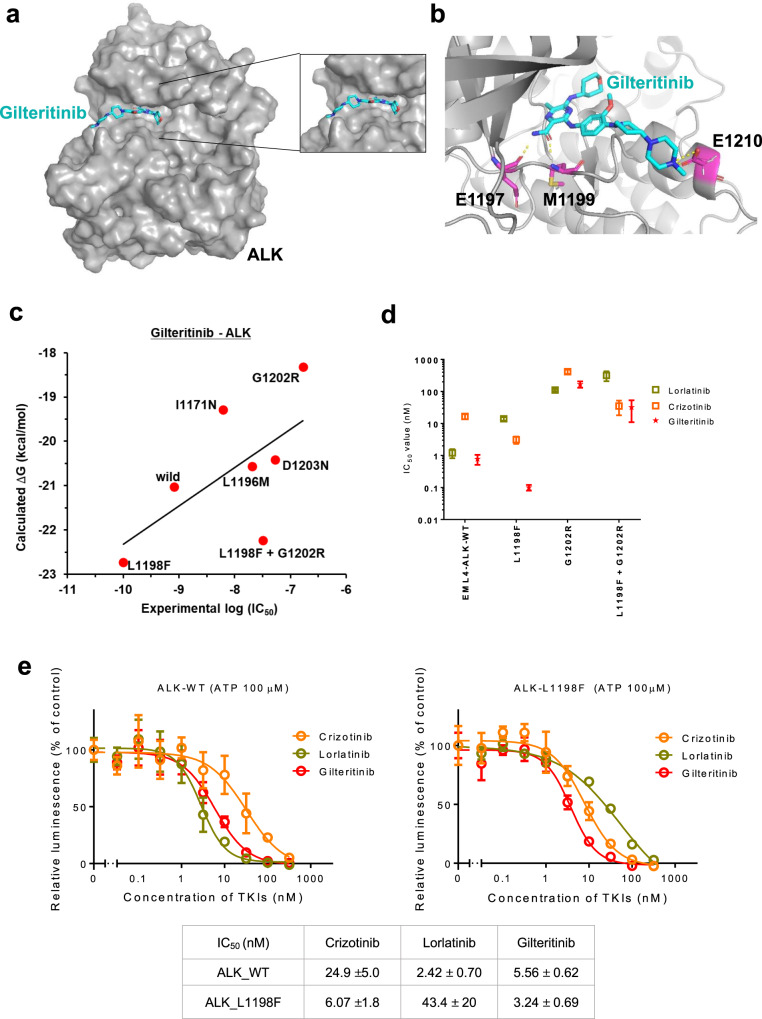
Fig. 6. Structure model of the ALK-gilteritinib complex and the predicted binding affinity for ALK mutants.
a The ALK–gilteritinib complex structure predicted by the molecular docking and molecular dynamic (MD) simulation. The mean stable structure of the ALK–gilteritinib complex was extracted from 50 ns × 5 MD simulations, and is represented by surface (ALK) and stick (gilteritinib: C, light blue; N, blue; O, red) models. In the structure model, gilteritinib fits into the ATP-binding pocket in ALK without any steric crushes by overview (left) and zoom-in of the ATP-binding pocket (right). b The gilteritinib-binding mode in the ATP-binding pocket in ALK. The protein backbone is represented by a gray ribbon diagram. E1197, M1199, and E1210 are colored with magenta, and their side chains were depicted as sticks (C, magenta; N, blue; O, red), respectively. Hydrogen bonds between these residues and gilteritinib are shown by dashed yellow lines. c The binding free energy (ΔG) of gilteritinib to wild-type (WT) or each resistant mutant is plotted against experimental IC50 of the corresponding Ba/F3 mutant. These ΔG values are calculated by MP-CAFEE. The linear association between ΔG and experimental IC50 was calculated by Pearson’s product–moment correlation coefficient (R = 0.627). d IC50 calculated from the cell viability assay of WT, L1198F, G1202R and L1198F + G1202R compound mutation-expressing Ba/F3 cells. Cells were treated with crizotinib, lorlatinib, or gilteritinib for 72 h. N = 3 independent samples examined over three independent experiments and data presented as mean values ± SD. e The evaluation of the sensitization activity of gilteritinib in the in vitro kinase assay using the ADP-Glo assay kit. IC50 value calculated at an ATP concentration of 100 μM suggested the better affinity of gilteritinib to ALK-L1198F than to wild-type ALK. N = 3 independent samples examined over three independent experiments and representative experiment data are presented as mean values ± SD.