Figure 1.
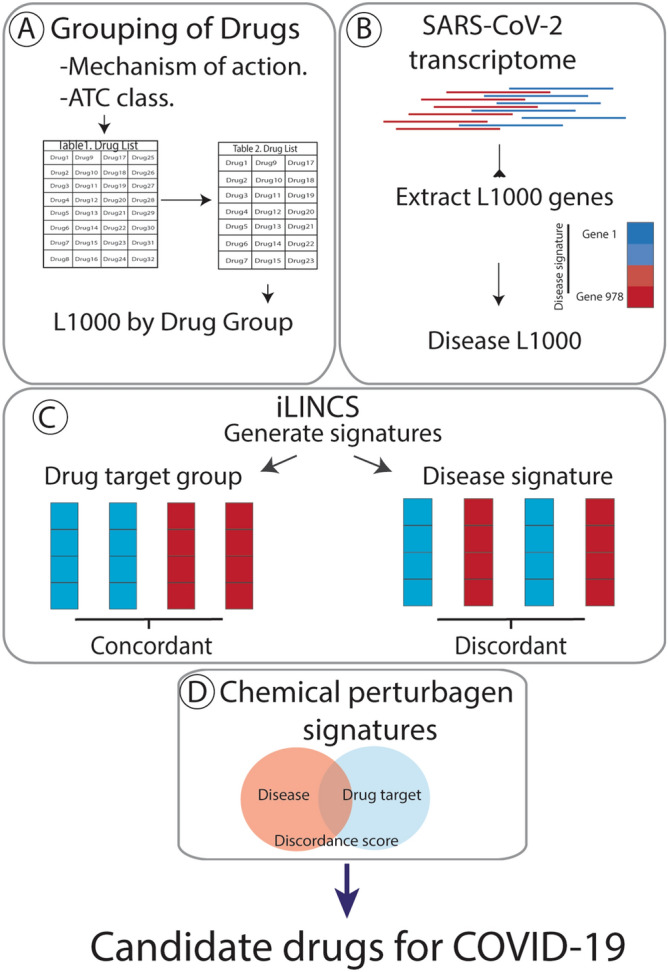
Overview of the workflow to identify candidate repurposable drugs to combat COVID-19. (A) Drugs that are currently in use to treat coronavirus and putative COVID-19 treatments were clustered based on mechanism of action and ATC class. (B) Gene expression data of the 978 genes that comprise the Library of Integrated Network-Based Cellular Signature (iLINCS) L1000 genes were extracted from severe acute respiratory syndrome coronavirus 2020 (SARS-CoV-2) (GSE147507) transcriptomic datasets. (C) Consensus iLINCS gene signatures were generated for drug groupings and disease. (D) Connectivity analysis was conducted and a list of chemical perturbagens that are concordant (≥ 0.321 concordance) to the drug target grouping signatures or discordant (≤ − 0.321 discordance) to the disease signatures was generated. Chemical perturbagens are filtered and curated to identify top candidate repurposable drugs.
