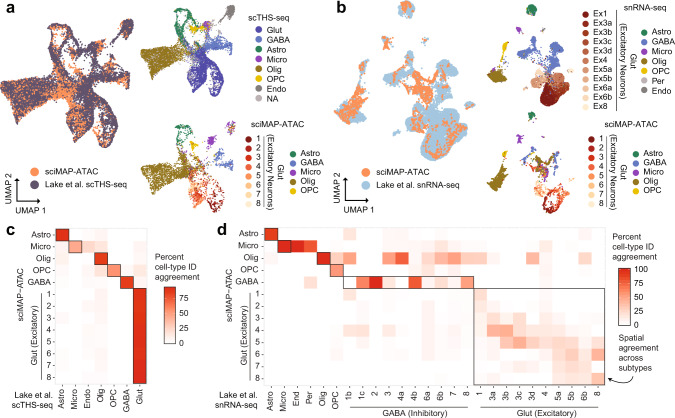
Fig. 5. Integration of sciMAP-ATAC with snRNA-seq and scTHS-seq human VISp datasets.
a Co-embedding of sciMAP-ATAC and scTHS-seq cell profiles from Lake et al.29 using Signac75 in a joint UMAP. Top right shows only scTHS-seq cells colored by cell type identified in Lake et al.29 and bottom shows sciMAP-ATAC cells colored by our called cell types as in Fig. 4c, except for glutamatergic neurons which are colored by spatial positions 1–8 (Glut glutamatergic (excitatory) neurons, GABA GABAergic (inhibitory) neurons, Astro astrocytes, Micro microglia, Olig oligodendrocytes, OPC oligodendrocyte precursor cells, Endo endothelial cells, NA not applicable—no cell type provided). b Co-embedding of sciMAP-ATAC and snRNA-seq transcriptional profiles from Lake et al.29 using Signac. Top right shows only snRNA-seq cells. Abbreviations as in a, but with the addition of Per = pericytes, and glutamatergic (excitatory) neurons (Ex) are colored by subtype identified in Lake et al.29. Bottom right shows only sciMAP-ATAC cells, with glutamatergic neurons colored by spatial position 1–8. c Confusion matrix representing the percent agreement in predicting the cell type of a cell from one dataset using the other between sciMAP-ATAC and scTHS-seq cells. d As in c, but between sciMAP-ATAC and snRNA-seq. Spatial agreement between excitatory neuron subtypes identified in the snRNA-seq data correspond to the spatial positioning of cells within our sciMAP-ATAC dataset. Source data are provided as a Source data file.