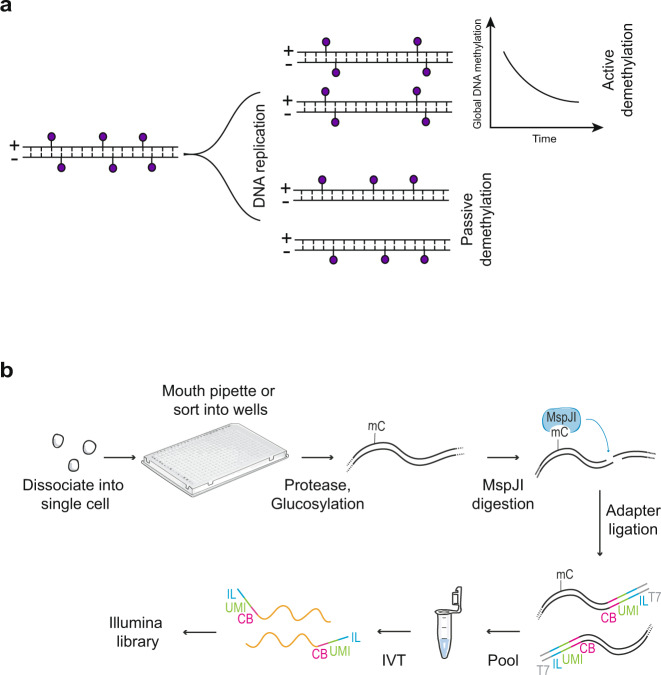
Fig. 1. Schematic of scMspJI-seq.
a DNA methylation maintenance can be probed using strand-specific quantification of 5mC in single cells. Cells displaying symmetric levels of 5mCpG on both DNA strands of a chromosome coupled with a global temporal loss of 5mCpG indicates active demethylation, whereas loss of methylation maintenance with asymmetric levels of 5mCpG between the two DNA strands indicates passive demethylation. b Single cells isolated by FACS or manual pipetting are deposited into 384-well plates and lysed. Following protease treatment to strip off chromatin and blocking of 5hmC sites by glucosylation, MspJI is used to recognize 5mC sites and cut gDNA 16 bp downstream of the methylated cytosine. After ligating double-stranded adapters—containing a cell-specific barcode (CB, pink), a random 3 bp unique molecule identifier to label individual 5mC sites on different alleles (UMI, green), 5′ Illumina adapter (IL, blue) and T7 promoter (T7, gray)—to the fragmented gDNA, molecules from all single cells are pooled and amplified by in vitro transcription. The amplified RNA molecules are used to prepare scMspJI-seq libraries and sequenced on an Illumina platform.