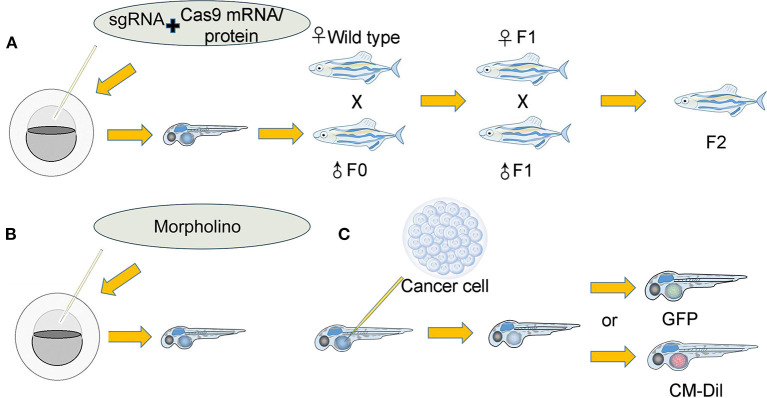
Figure 2.
Establishment of the tumor microenvironment in the zebrafish xenograft model. (A): (I) A single-guide RNA (sgRNA) was coinjected along with Cas9 mRNA/protein into either the yolk or cell of single-cell stage embryos. The coinjected embryos grew into founder fish. (II) The founder fish were then outcrossed to wild-type siblings to generate heterozygous F1 fish. Fish lacking the correct genotype (as identified by fluorescence PCR and sequencing) were eliminated. (III) The F1 fish carrying the correct mutations were then crossed to generate F2 progeny. F2 offspring will provide an optimal tumor microenvironment for researchers. (B): Morpholino knockdown of specific genes in zebrafish embryos directly provides the tumor microenvironment. (C): (I) Cancer cells that were either expressing GFP or labeled with other fluorescent dyes were implanted into zebrafish. (II) Various indexes of cancer cells were observed and recorded.