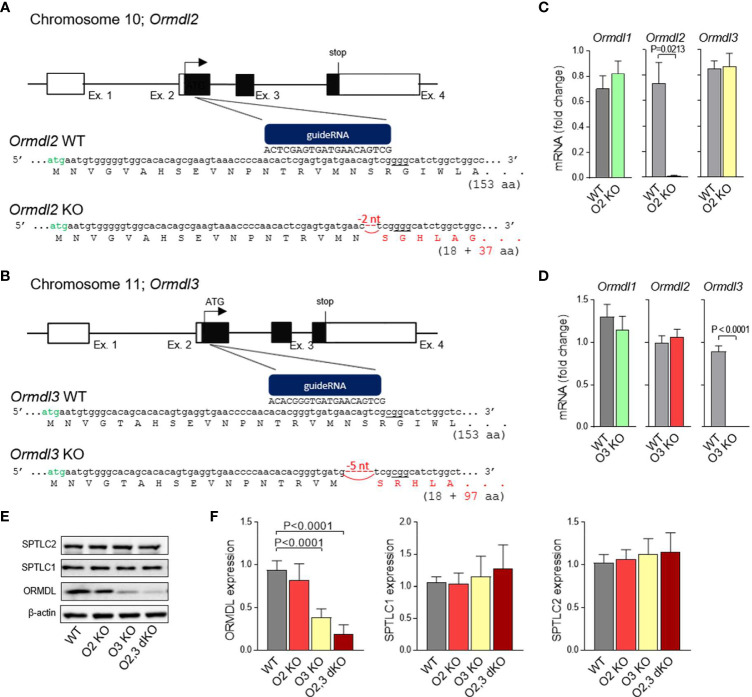
Figure 1.
Production and initial characterization of ORMDL2 KO, ORMDL3 KO, and ORMDL2,3 dKO. (A, B) Strategy to generate ORMDL2 KO (O2 KO) and ORMDL3 KO (O3 KO) mice using the CRISPR/Cas9 approach. Specific guideRNAs were prepared to target the Ormdl2 and 3 within exon 2 (sites underlined) downstream of a second ATG codon (in green). Founders with 2-nucleotide or 5-nucleotide deletion leading to a frame shift in a coding sequence of the Ormdl2 and Ormdl3 gene, respectively, were used to establish the ORMDL2 or ORMDL3 KO mice. (C) RT-qPCR quantification of mRNAs encoding ORMDL1, ORMDL2, and ORMDL3 proteins in mast cells isolated from WT mice (n = 4) and O2 KO mice (n = 4). (D) RT-qPCR quantification of mRNAs encoding ORMDL1, ORMDL2, and ORMDL3 proteins in mast cells isolated from WT mice (n = 8) and O3 KO mice (n = 11). ORMDL2 KO and ORMDL3 KO mice were further crossed to produce ORMDL2,3 double KO (O2,3 dKO) mice. (E) Representative immunoblot of the lysates from BMMCs isolated from WT or O2 KO, O3 KO, and O2,3 dKO mice was developed with pan-ORMDL-specific antibody and the antibodies recognizing long chain subunits of the serine palmytoil transferase (SPT) complex. (F) Statistical analysis of the data presented in (E); WT (n = 6), O2 KO (n = 6), O3 KO (n = 6), and O2,3 dKO (n = 6). Quantitative data are mean ± s.e.m., calculated from n, which shows numbers of biological replicates. P values were determined by unpaired two-sided Student’s t-test in (C, D), or by one-way ANOVA with Tukey’s post hoc test in (F).