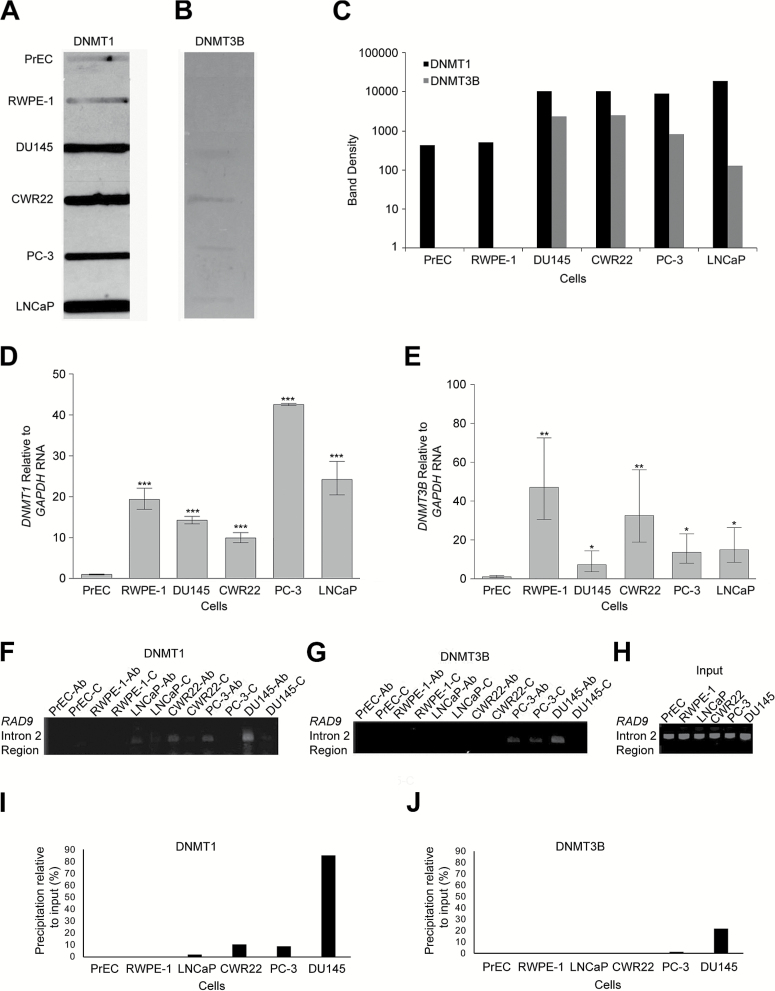
Figure 1.
DNMT1 and DNMT3B DNA methyltransferase levels are high in multiple prostate cancer cell lines, relative to non-cancer prostate cells. However, both enzymes bind the transcription suppressor region of RAD9 intron 2 primarily in DU145, the only cells tested where methylation regulates RAD9 expression. The in vivo complex of methylation (ICM) assay was used to measure complexes of (A) DNMT1 and (B) DNMT3B with aza-dC adducts in genomic DNA, formed in vivo, within PrEC, RWPE-1, DU145, CWR22, PC-3 and LNCaP cells. (C) Slot blot band densities were quantified. ICM was performed three times and a representative slot blot as well as slot band density profiles are presented. Quantitative RT–PCR was used to measure (D) DNMT1 and (E) DNMT3B RNA abundance relative to GAPDH in each of the cell lines, and values were normalized to results from PrEC. Experiments were performed in triplicate, and average values are presented. Bars indicate standard error of the mean. p values: ‘***’, ≤ 0.001; 0.001< ‘**’ ≤ 0.01; 0.01< ‘*’ ≤ 0.05. Chromatin immunoprecipitation (ChIP) was used to determine binding of (F) DNMT1 and (G) DNMT3B to the transcription suppressor domain of RAD9 intron 2. Studies were performed with PrEC, RWPE-1, LNCaP, CWR22, PC-3 and DU145 cells. ‘Ab’ indicates that DNMT1 or DNMT3B antibodies were used for the assay, and ‘C’ indicates use of IgG control antibodies. (H) Input DNA is indicated. Band densities derived from ChIP with either (I) DNMT1 or (J) DNMT3B antibodies, minus background ChIP using IgG antibodies, relative to input DNA band densities are presented. Representative data from three independent trials are shown.