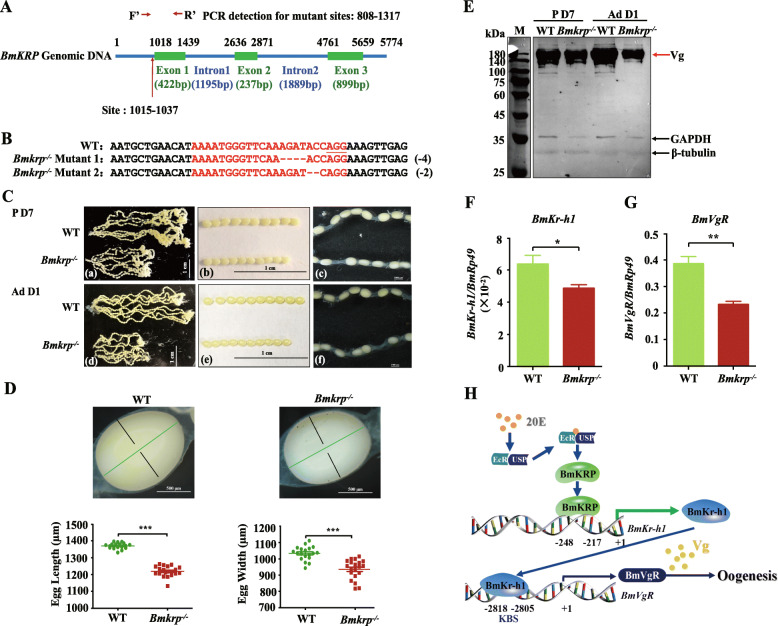
Fig. 5.
Effects of knockout of BmKRP by CRISPR/Cas9-mediated genome editing on ovarian development and oogenesis. A Schematic diagram of the sgRNA-target site. The green boxes indicate the three exons of BmKRP, and the blue line indicates the gene locus. The sgRNA targeting site was located on the sense strand of exon 1. F′and R′ were used for annealing the upstream and downstream regions of the targeting site. B Two mutant types of deletion screened from homozygous mutant silkworms in G3 generation. The wild-type sequence is shown at the top with the sgRNA target sites marked in red and PAM sequences is underlined. In the mutant sequences, deletions are shown as dashes. C Phenotypes of ovaries and oocytes of 7-day-old pupae (a–c) and 1-day-old adults (d–f) in G3 generation. D Effects of knocking down BmKRP on oocyte length and width of 7-day-old pupae. Green line represents the length and black line represents width. E Western blot analysis of BmVg protein in the oocyte. A total of 15 μg protein from the oocytes was loaded per lane and probed with anti-BmVg, anti-GAPDH, and anti-β-tubulin antibodies. qRT-PCR analysis of BmKr-h1 (F) and BmVgR (G) mRNA in the oocytes of 7-day-old pupae. Rp49 amplified from the same RNA samples was used as an internal control. H Schematic representation of the 20E-induced and BmKRP mediated expression of BmKr-h1 and its physiological function in oogenesis. WT, wild-type. The data in F and G are means ± SEM (n = 3) and the individual data values are shown in Additional file 2. The significance of the differences between the treatment and control was statistically analyzed at p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) using t test