Extended Data Fig. 3 |. Cryo-EM data processing for FPN-Fab45D8.
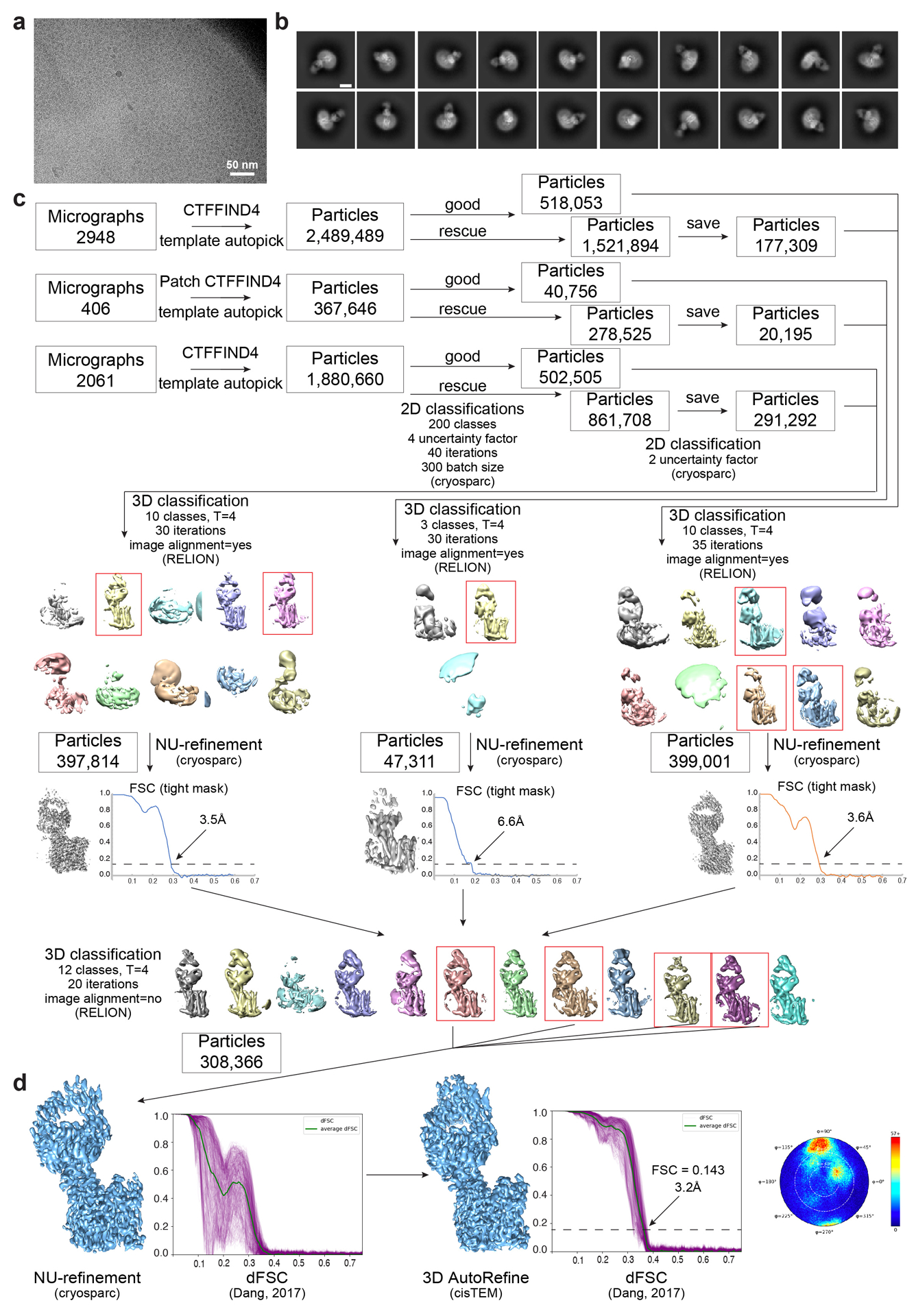
a, Representative motion-corrected micrograph collected on the Titan Krios showing monodisperse FPN-Fab45D8 nanodisc particles. Four different FPN-Fab45D8 samples were imaged on GO-coated Quantifoil grids times with similar results. b, Examples of “good” 2D class averages that were used in 3D classification. Similar quality class averages were produced when processing six unique subsets of the final particles used. Scale bar is 50 Å. c, Flowchart showing image processing pipeline for FPN-Fab45D8. Initial processing, through 2D classification, was performed in cryoSPARC. Particles were then transferred, using csparc2star.py, to RELION for 3D classification, then to cryoSPARC for a nonuniform refinement, and finally to cisTEM for refinement. The number of particles moving into each step are noted. d, Final refinements from cryosparc and cisTEM beside their directional FSC curves calculated using dfsc.0.0.1.py. Angular distribution plot from cisTEM is shown.
