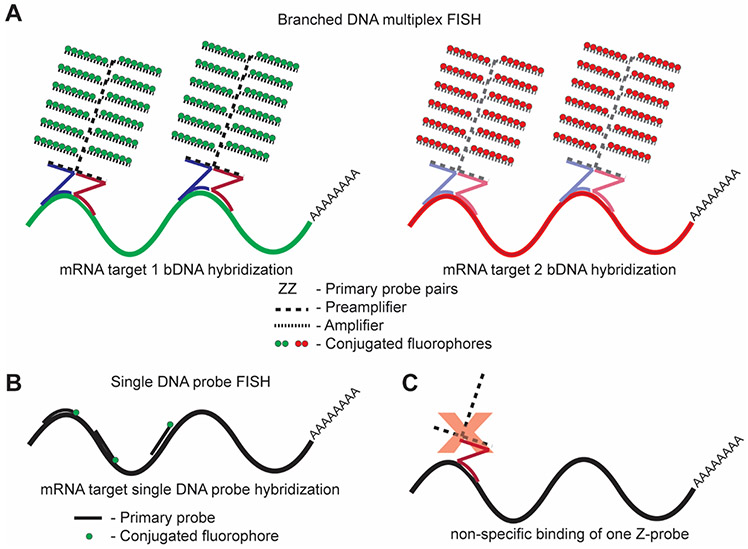
Fig. 1. Overview of the branched DNA approach.
(A) In bDNA assays, pairs of primary probes (ZZ) identify and hybridize to a specific gene product of interest. Preamplifiers and amplifiers then bind the probe pairs to form tree-like structures. Fluorescent label probes attach to respective amplifier “branches” to give up to 100× higher signals than approaches illustrated in (B) under equivalent imaging conditions. Both ViewRNA and RNAscope follow the same general cascade of hybridization events. Label probes for ViewRNA are predetermined at the time of probe set design. Label probe combinations for RNAscope are determined at the time of the experiment according to Table 1. (B) Alternative FISH approaches include the direct hybridization of several short oligonucleotide probes with a single fluorophore, which require long transcripts to ensure high fluorescence intensity for detection [38, 41]. While direct fluorophore conjugation does not allow for signal-to-noise ratios as high as those afforded by bDNA assays, as described by Titlow et al., a benefit of this method is the capability of viewing protein localization in parallel with RNA localization [38]. (C) An advantage of using a bDNA approach for RNA in situ hybridization is the high specificity of probe pairs compared to individual oligonucleotide probes. If only one member of a probe pair hybridizes to an off-target RNA sequence, the preamplifier cannot bind, and therefore no fluorescent signal will result.