Figure 6: FGFR-ERLIN2 fusion in PAX265.
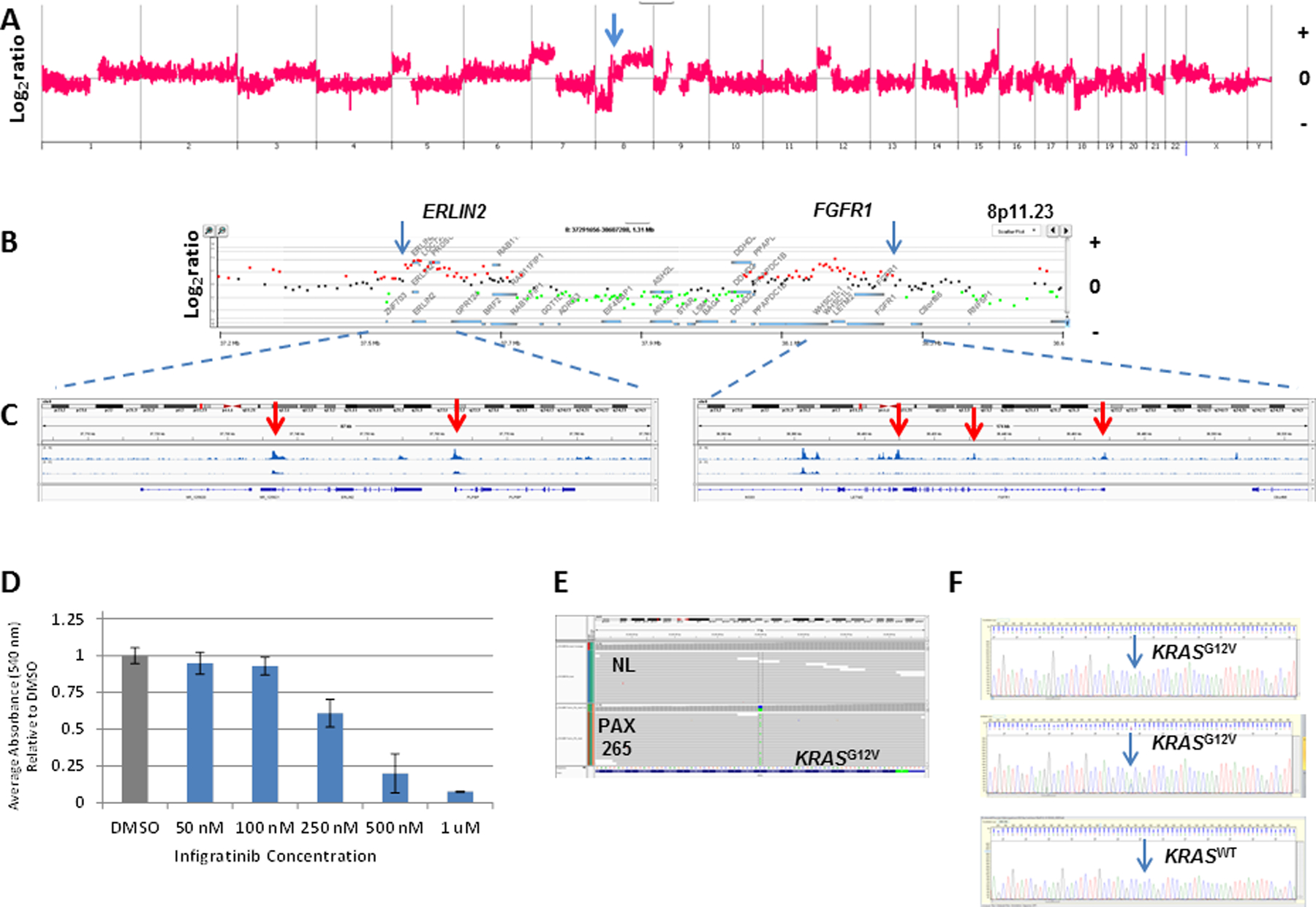
A) Whole genome and; B) locus specific analysis of CNV gains targeting ERLIN2 and FGFR1. C) IGV views of ATAC-seq profiles of ERLIN2 (left panel) and FGFR1 (right panel). Red arrows denote open chromatin peaks in each gene. D) Dose response profile of PAX265 organoids to Infigratinib. Organoids were seeded in 96-well plate coated with Matrigel (5,000 cells /well) in organoid media. After 2–3 days of growth, fresh organoid media and DMSO or drug (10 wells/condition) was added. Fresh media and drug was added every 2–3 days. MTT assay were done on day 7 to measure proliferation. The absorbance listed for each drug is the average of the 10 wells per condition. Experiment was performed in triplicate with an average IC50 near 250 nM. E) IGV view of KRASG12V mutation in flow sorted PAX265. F) Single cell sequencing of KRAS exon 2 in flow sorted PAX265 derived organoids. Sanger sequencing of single nuclei sorted from 50 individual organoids identified 19 homozygous (top) and 15 heterozygous (middle) KRASG12V variants. The other 16 were KRASG12G wild type (bottom).
