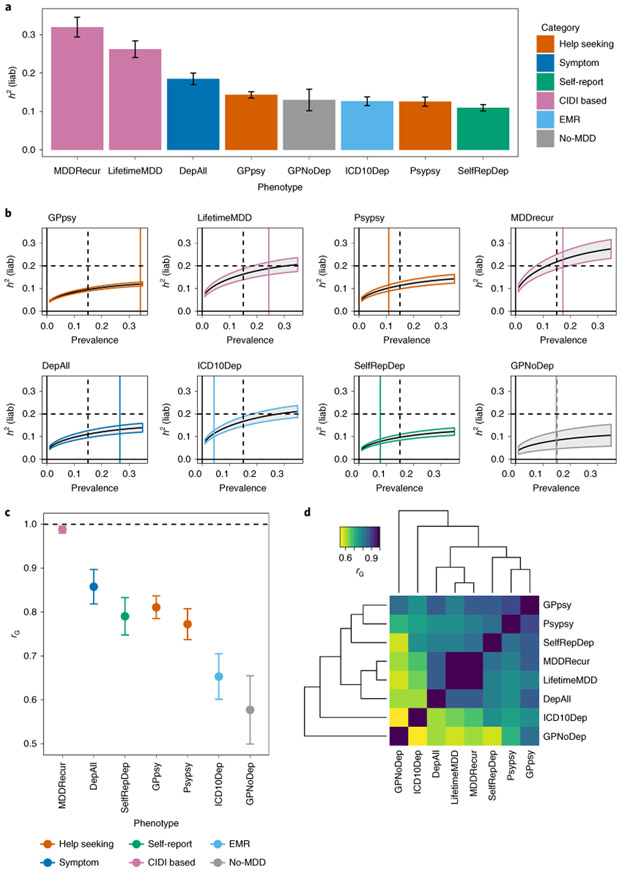
Fig. 3 ∣. SNP heritability and genetic correlation estimates among definitions of MDD in UK Biobank.
a, h2SNP estimates from PCGC18 on each of the definitions of MDD in the UK Biobank (Methods). h2SNP (represented as h2(liab)) was converted to the liability scale40,63 using the observed prevalence of each definition of depression in the UK Biobank as both population and sample prevalence (Supplementary Table 4). Error bars show the s.e. of the estimates. b, h2SNP estimates of definitions of MDD in the UK Biobank from LDSC using logistic regression summary statistics on all SNPs with minor allele frequency (MAF) > 5% (Methods), transformed to the liability scale assuming a range of population case prevalence values, from 0 to 0.5. We do not show results for case prevalence from 0.5 to 1, as they would mirror those from 0 to 0.5, with shaded area representing the s.e. of the estimates. We indicate with a black vertical dashed line the population prevalence of 0.15, used in PGC1-MDD; a colored vertical line shows the population prevalence of each definition of depression in the UK Biobank. We also indicate with a black horizontal dashed line the arbitrary liability-scale h2SNP of 0.2, previously estimated for MDD in PGC1-MDD. Using this, we show that at no prevalence would minimal phenotyping-defined depression such as GPpsy (help-seeking definition) reach this estimate. c, Genetic correlation ‘rG’ between CIDI-based LifetimeMDD and all other definitions of MDD in the UK Biobank, estimated using PCGC. Error bars show the s.e. of the estimates. d, Pairwise rG between all definitions of depression in the UK Biobank, also detailed in Supplementary Table 15.