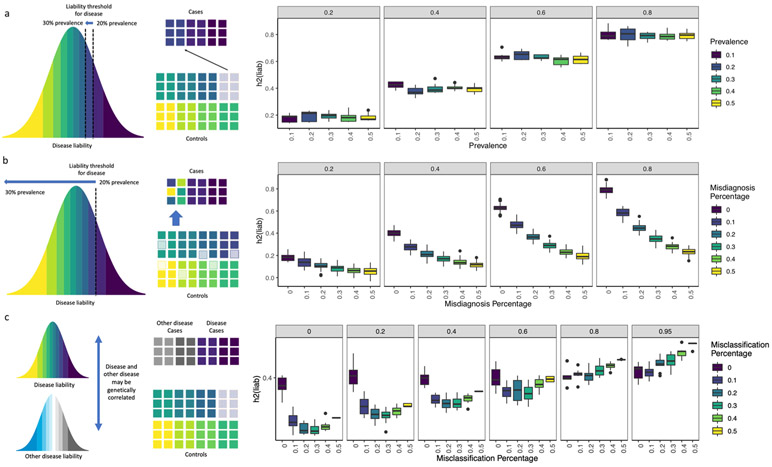
Extended Data Fig. 1 ∣. Simulations of misdiagnosis and misclassification.
a-c, Each boxplot show h2SNP estimates from 10 simulated phenotypes, with upper and lower boundaries of boxes represent the first to third quartiles of all estimates, and the whiskers extends to 1.5 times the interquartile range of the estimates. a, This figure shows that liability scale h2SNP does not change with shifting of liability threshold Ki∈{0.1, 0.2, 0.3, 0.4, 0.5} for simulated heritabilities ∈ {0.2, 0.4, 0.6, 0.8}. b, The figure shows that liability scale h2SNP is deflated with increasing percentage of controls being misdiagnosed as cases, when prevalence of diagnosed cases is kept constant at Ki=0.2, for simulated heritabilities ∈ {0.2, 0.4, 0.6, 0.8}. c, This figure shows liability scale h2SNP is deflated with increasing percentage of misclassification of cases of “other” disease as cases of focal disease, if rG between the two diseases are moderate to low, for simulated , for each of which all cases at prevalence Ki,1=0.2 are correctly identified as cases.