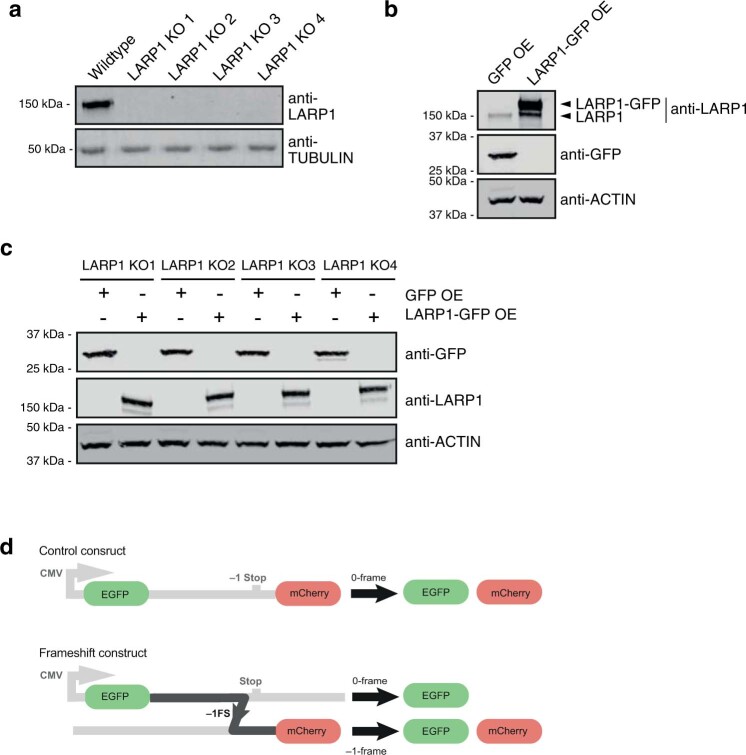
Extended Data Fig. 4. Functional validation of SARS-CoV-2 RNA binders.
a, Western blot of WT HEK293 cells and four different HEK293 LARP1 knockout (KO) cell lines generated with CRISPR-Cas9 (see Methods). Expression of LARP1 was evaluated relative to Tubulin. b, Western blot of HEK293 cells transiently overexpressing (OE) GFP or LARP1-GFP proteins at 48 h post transfection. Arrows indicate endogenous LARP1 proteins and GFP-tagged LARP1. c, Western blot of four different HEK293 LARP1 knockout cell lines transiently transfected with plasmids encoding GFP or LARP1-GFP proteins at 48 h post transfection. Experiments were repeated at least two times. d, Schematic of dual-fluorescence translation reporter to quantify ribosomal frameshifting efficiency. The depicted control construct contains enhanced GFP (eGFP) and mCherry in an in-frame orientation, leading to the production of both fluorescent proteins separated by a self-cleaving 2A peptide when the 0 reading frame is translated. In the frameshift construct depicted below, eGFP and mCherry are separated by an in-frame stop codon, preventing the production of mCherry when the 0 reading frame is translated. −1FS leads to the production of eGFP and mCherry and the ratio between both fluorescent proteins is a direct measure of frameshifting efficacy. −1FS: –1 ribosomal frameshifting.