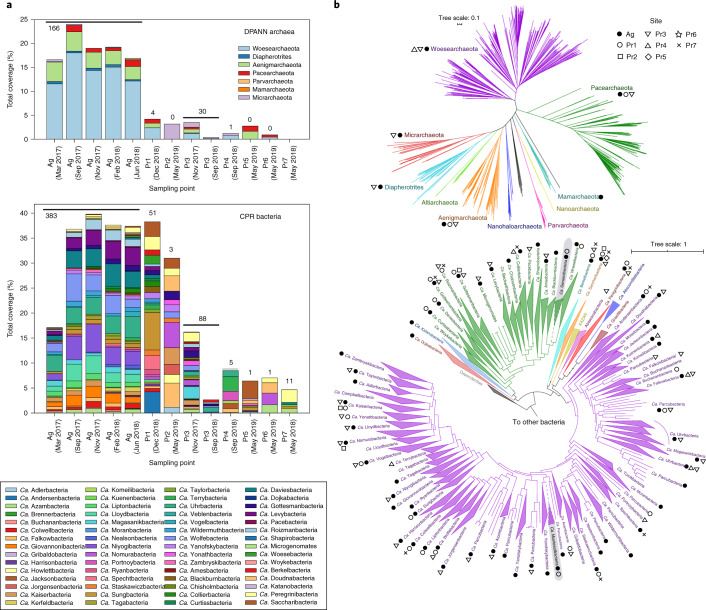
Fig. 2. Distribution of CPR and DPANN organisms across groundwater sites.
a, Abundances (relative coverage of scaffolds containing rpS3 marker genes) of phylum-level lineages within DPANN (above) and CPR (below) in all sites. The numbers above the bars indicate the number of draft-quality (>70% complete, <10% contamination) dereplicated DPANN or CPR genomes that were recovered from metagenomic reads. b, Maximum likelihood phylogenetic tree of the DPANN radiation (top), based on 14 concatenated ribosomal proteins, and of the CPR (bottom), based on 15 concatenated ribosome proteins. Phylum-level lineages within the CPR (as previously defined2) are collapsed. Markers next to each lineage indicate the groundwater sites where at least one representative genome from that lineage was recovered. New CPR lineages ‘Candidatus Genascibacteria’ (within the Microgenomates superphylum in green) and ‘Candidatus Montesolbacteria’ (within the Parcubacteria superphylum in purple) are highlighted in grey.