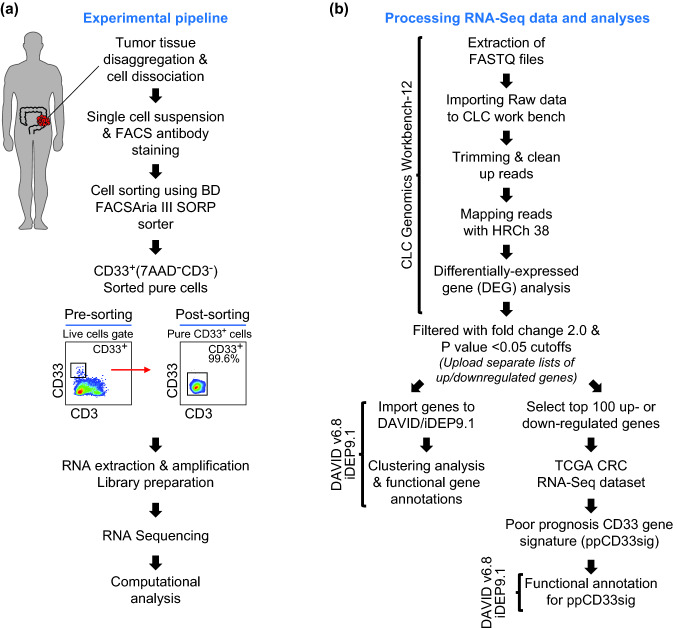
Fig. 1.
Schematic representation of study design. Tumor tissue specimens from 13 CRC patients were used to sort highly pure CD33high myeloid cells. Libraries were generated from sorted cells for RNA-Seq. Different bioinformatics tools were utilized for analyses and visualization of RNA-Seq data. The experimental flowchart of this study is shown in (a), and the workflow for RNA-Seq data pre-processing and analysis using various bioinformatics tools is shown in (b)