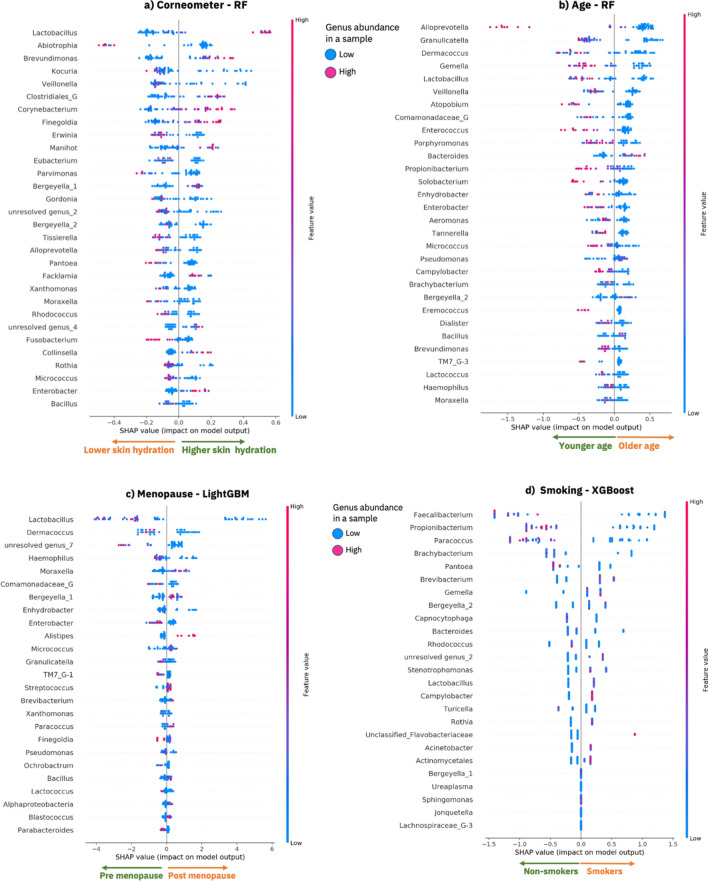
Figure 2.
Model explanations for the 62 first samples taken from the Canada cohort. SHAP summary dot plot as computed by SHAP using the best optimized ML model that has been trained on 80% of the training data. Each plot provides an overview of which features are most important for a model and visualizes how the value of each feature (i.e., the genus abundance in the samples) contributes, either positively or negatively, to the prediction of phenotypic values; (a) lower or higher corneometer measurements, (b) lower or higher values of age, (c) pre-menopausal or post-menopausal status and (d) non-smokers or smokers. The features are sorted by the sum of the absolute SHAP values over all the samples in the training dataset. Each dot is a sample, and its color represents a feature value (i.e., genus abundance) for the sample. Red dots are samples for which a genus (row) is enriched, while blue dots are samples for which a genus is lower in abundance. Clusters of red samples on the right side of the x-axis means that the genus is abundant in those samples and it is contributing to the prediction of a higher phenotypic value (indicated by the x-axis annotation of arrows pointing right). Clusters of red samples (dots) on the left side of the y-axis means that the genus is enriched for those samples and it is contributing to the prediction of a lower phenotypic value for those samples (indicated by the x-axis annotation of arrows pointing left). This image was created using SHAP20 version 0.34.0 (https://github.com/slundberg/shap).