Figure 3.
Transcriptional regulation of iTreg cells by Nr4a factors
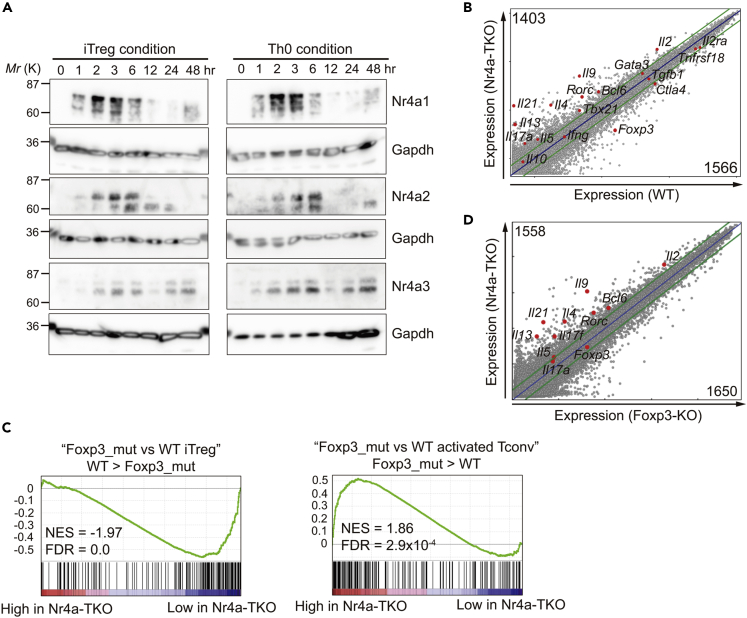
(A) Western blot analysis showing the time course of Nr4a family protein expression under indicated culture conditions.
(B) mRNA expression profiles of wild-type (WT, horizontal axis) and Nr4a triple-knockout (TKO, vertical axis) induced Treg (iTreg) cells at 24 hr, analyzed by microarray. Selected Th- and Treg-regulatory genes are highlighted as red dots. A number of genes differentially expressed (|log2| > 1) are shown.
(C) GSEA enrichment plot for the indicated MSigDB Hallmarks against the microarray data of WT iTreg at 24 hr vs. Nr4a-TKO iTreg at 24 hr. NES, normalized enrichment score; FDR, false discovery rate.
(D) Expression profiles of Foxp3-knockout (KO, horizontal axis) and Nr4a-TKO (vertical axis) iTreg cells at 24 hr, analyzed by microarray. Selected Th- and Treg-regulatory genes are highlighted as red dots. A number of genes differentially expressed (|log2| > 1) are shown.