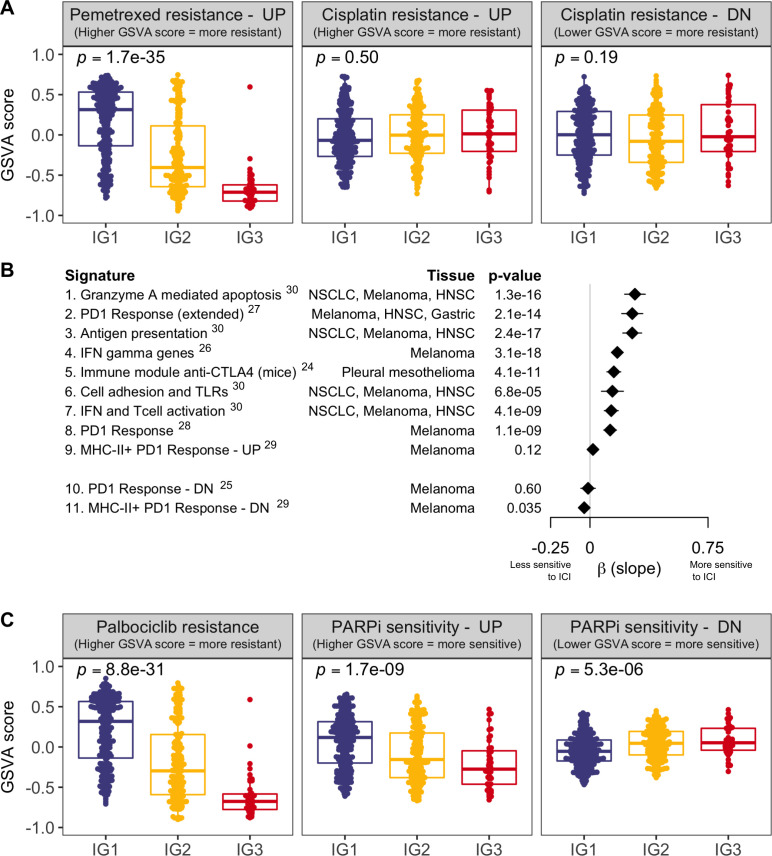
Figure 5.
Assessment of gene expression signatures predictive of benefit or resistance to multiple treatments. (A) Enrichment assessment for signatures of resistance to currently established first-line chemotherapy in patients with MPM (pemetrexed—left panel, cisplatin—mid and right panels). Each MPM tumor is represented with a dot, and GSVA scores (y-axis) indicate upregulation or downregulation of the signature in each patient (as indicated in each plot title). Signature of resistance to cisplatin is divided into two subsignatures of upregulated and downregulated genes in cisplatin resistant cells. (B) Assessment of response to multiple ICI therapies. Each line represents a different ICI predictive signature. Signatures 1–9 are upregulated in ICI responders, while signatures 10 and 11 are downregulated in ICI responders, based on the literature. For each signature, we represent the β coefficient and the 95% CI of the linear model of the GSVA scores across the three immune groups. Positive β values indicate increasing GSVA scores (IG1<IG2<IG3) and negative β values indicate decreasing GSVA scores (IG1>IG2>IG3). Superscripts in immunotherapy signatures correspond to bibliographic references (C) Analogously to panel A, assessment for signatures of resistance or sensitivity to targeted therapy (palbociclib—left panel, PARP inhibitors—mid and right panels). Signature of sensitivity to PARP inhibitors is divided into two subsignatures of upregulated and downregulated genes in PARP inhibitors sensitive cells. GSVA, gene set variation analysis; HNSC, head and neck squamous carcinoma; ICI, immune checkpoint inhibitor; IFN, interferon; MHC, major histocompatibility complex; NSCLC, non-small cell lung carcinoma; PARP, poly ADP ribose polymerase; PD1, programmed cell death protein 1; TLR, toll-like receptor; UP/DN, up/down-regulated gene signature in either sensitive or resistant cells for the specified treatment.