Figure 4.
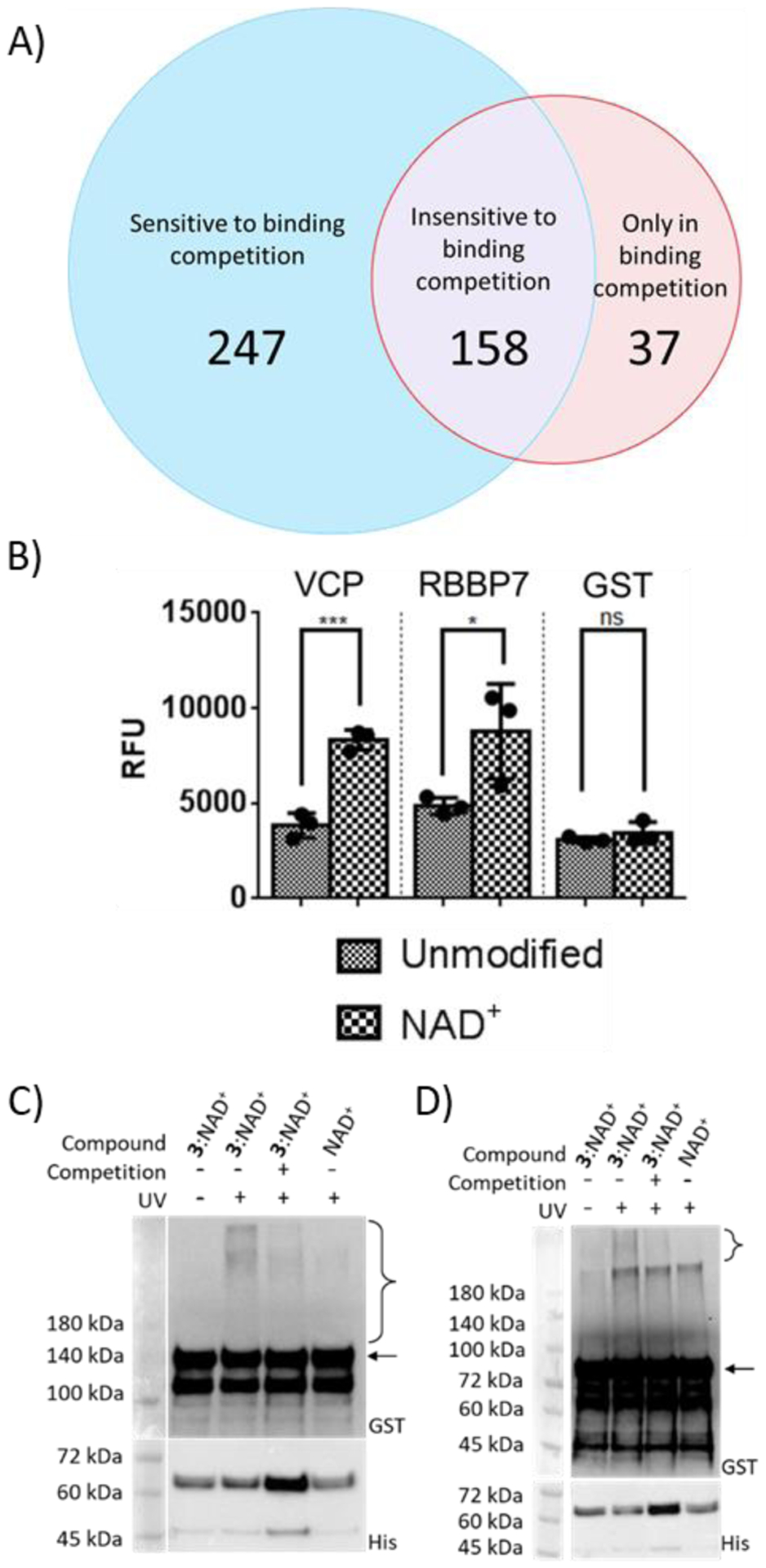
Verification of novel PARylation-dependent interacting proteins. (A) Venn diagram of protein hits identified from photocrosslinking in cell lysates. (B) Binding of GST-VCP and GST-RBBP7 for unmodified PARP1 and automodified PARP1 by NAD+. Unmodified PARP1 and NAD+-modified PARP1 were coated on ELISA plates for binding analysis with two identified protein hits. ns: not significant, *P < 0.05, *** P < 0.001 by one-tailed unpaired t-test. Error bars represent standard deviation of three replicates. RFU: relative fluorescence unit. (C) and (D) Photocrosslinking of automodified PARP1 with (C) VCP and (D) RBBP7. PARP1 PARylated by NAD+ or a mixture of 3:NAD+ (molar ratio 1:1) was incubated with GST-VCP or GST-RBBP7 without and with 365-nm UV and NAD+-modified PARP1 for binding competition, followed by immunoblot analysis using an anti-GST antibody. Expected sizes for VCP and RBBP7 are indicated by the arrow for (C) and (D) respectively. Lower panels: PARP1 loading controls detected using an anti-His6 antibody. The observed bands between 45 and 72 kDa were proteolyzed PARP1 formed during auto-modification reactions. Brackets indicate regions of crosslinked protein complexes.
