Figure 1.
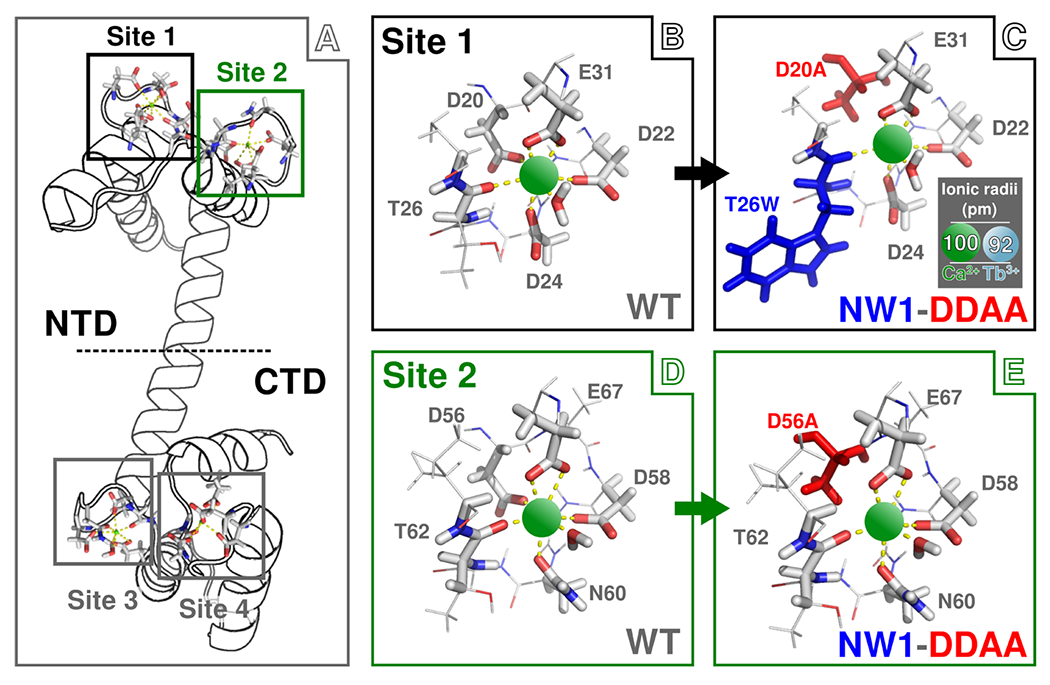
Schematic representation of the wild-type (WT) and mutant N-CaMs used in this study bound to Ca2+. (A) Structure of full-length WT CaM. Binding site (BS) locations are highlighted by black (BS 1), green (BS 2), and gray (BS 3 and BS 4) boxes. For this work, we use only the sequence of N-terminal domain (NTD) CaM, which contains BS 1 and BS 2 and appears above the dashed horizontal line. (B) Structure of WT BS 1. Ca2+ ions are shown as green spheres connected by yellow dashed lines to carboxylate and carbonyl groups responsible for coordination within the binding site. (C) Structure of BS 1 with both NW1 (colored blue) and DDAA (colored red) mutations. (D) Structure of WT BS 2. (E) Structure of BS 2 with both the NW1 and DDAA mutations. Note that the NW1 mutation affects only BS 1; thus, BS 2 of the NW1 mutant is the same as WT. The DDAA mutant involves mutation of both aspartate 20 in BS 1 and aspartate 56 in BS 2 to alanine; thus, the DDAA mutant involves aspartate-to-alanine mutations in both BS but no insertion of tryptophan in either site. The NW1-DDAA mutant combines both the NW1 and DDAA mutations. The inset in panel C shows the relative sizes of the Ca2+ and Tb3+ to scale with the BS structure. Note that these structures are shown only for illustration of point mutation; actual binding site structures differ and are both sequence-and ion-dependent.
