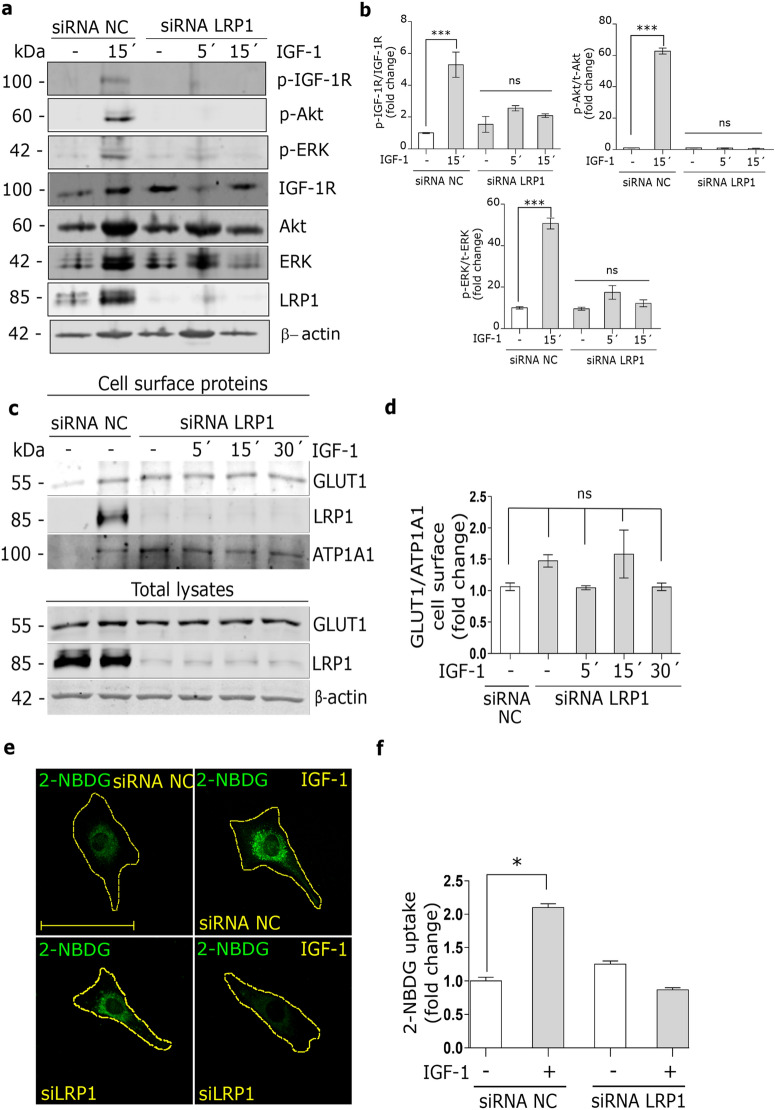
Figure 5.
IGF-1-induced GLUT1 traffic to PM and glucose uptake are dependent on LRP1 expression. (a) Western blot assay for the analysis of phosphorylated IGF-1R (p-IGF-1R; T1316), Akt (p-Akt; T308) and ERK 1/2 (p- ERK1/2; Thr202/Tyr204) in MIO-M1 cells treated or not with siRNA for LRP1 and then stimulated with IGF-1 10 nM for 5–15 min. Total IGF-1R, Akt, ERK1/2 and β-actin were used as loading control. (b) Densitometric quantification of Western blot data expressed as fold change respect to non-stimulated control (white bar). Values are expressed as mean ± SEM. ***p < 0.001 versus non-stimulated control. ns = non-significant differences. Three independent experiments in duplicate were performed (n = 6). (c) Biotin-labeling protein assay to measure expression of GLUT1 and LRP1 in the PM of MIO-M1 cells treated or not with siRNA for LRP1 and then stimulated with IGF-1 10 nM for 5–30 min. Biotin-labeled proteins were isolated with streptavidin-conjugated beads and then analyzed by Western blot. ATP1A1 and β-actin were used as protein loading controls. Line 1: control without biotin. (d) Densitometric quantification of Western blot data for surface GLUT1 related to ATP1A1 expressed as fold change respect to non-stimulated control (white bar). Values are expressed as mean ± SEM. ns = non-significant differences. Three independent experiments in duplicate were performed (n = 6). (e) Confocal microscopy in MIO-M1 cells treated with specific siRNA for LRP1 (siLRP1) and then stimulated with IGF-1 10 nM together with 2-NBDG 80 µM (green) for 30 min. Dotted line represents the cell shape. Images are representative of 20 cells per condition (n = 20). Scale bar = 10 μM. (f) Graph represents mean ± SEM of the fluorescence intensity of 2-NBDG per cell area expressed as fold change. *p < 0.05 versus non-stimulated control (white bars).