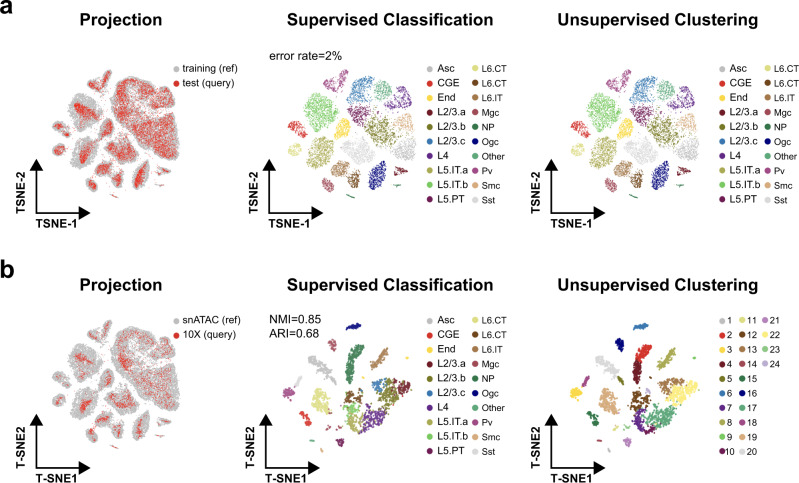
Fig. 6. SnapATAC enables supervised annotation of new scATAC-seq dataset using reference cell atlas.
a MOs snATAC-seq dataset is split into 80% and 20% as training and test dataset. A predictive model learned from the training dataset predicts cell types on the test dataset of high accuracy (left) as compared to the original cell-type labels (right). b A predictive model learned from the reference dataset—MOs (snATAC)—accurately predicts the cell types on a query dataset from mouse brain generated using a different technological platform, the 10× scATAC-seq. The t-SNE embedding is inferred from the reference cell atlas (left) or generated by SnapATAC in an unbiased manner from 10× mouse brain dataset (middle and right). Cells are visualized using t-SNE and are colored by the cell types predicted by supervised classification (middle) compared to the cluster labels defined using unsupervised clustering (right).