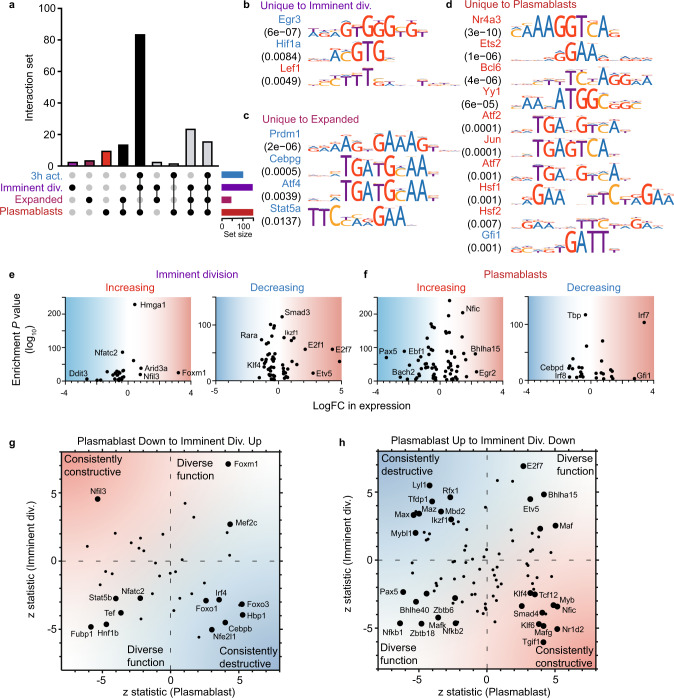
Fig. 4. Numerous transcription factors regulate B-cell differentiation via 3D genome organization.
a Upset plot showing shared and unique enriched motifs in DIP patterns that are exclusively increasing or decreasing at the transitions. The vertical bars indicate the number of motifs shared between the patterns and the black dots indicate the patterns shared. The horizontal bars denote the number of motifs enriched in the pattern. b Motifs enriched uniquely in the exclusively increasing or decreasing 10 h activation to imminent division DIP patterns ranked by adjusted p-value. The color of the motif indicates whether it was enriched in the increasing (red) or decreasing (blue) DIPs pattern. c Motifs enriched uniquely in the exclusively increasing or decreasing imminent division to expanded patterns ranked by adjusted p-value. d Motifs enriched uniquely in the exclusively increasing expanded to plasmablasts patterns ranked by adjusted p-value. e The adjusted p-value (−log10) of the motifs enriched in the exclusively increasing or decreasing 10 h activation to imminent division DIP patterns plotted as a function of logFC of expression at that transition. Blue shading represents decreased expression, red represents increased expression. f The adjusted p-value (−log10) of the motifs enriched in the exclusively increasing or decreasing expanded to plasmablasts DIP patterns plotted as a function of logFC of expression at that transition. Blue shading represents decreased expression, red represents increased expression. g The z-statistic of expression for the 10 h activation to imminent division transition plotted against the z-statistic of expression for the expanded to plasmablasts. The genes plotted correspond to motifs enriched in the exclusively increasing 10 h activation to imminent division DIP pattern and exclusively decreasing expanded to plasmablasts DIP pattern. Labeled genes are significant in both transitions (FDR < 0.05). Blue shading represents potentially destructive factors, red represents potentially constructive factors. h The z-statistic of expression for the 10 h activation to imminent division transition plotted against the z-statistic of expression for the expanded to plasmablasts. The genes plotted correspond to motifs enriched in the exclusively decreasing 10 h activation to imminent division DIP pattern and exclusively increasing expanded to plasmablasts DIP pattern. Labeled genes are significant in both transitions (FDR < 0.05). Blue shading represents potentially destructive factors, red represents potentially constructive factors. Source data are provided as a Source Data file.