Fig. 1.
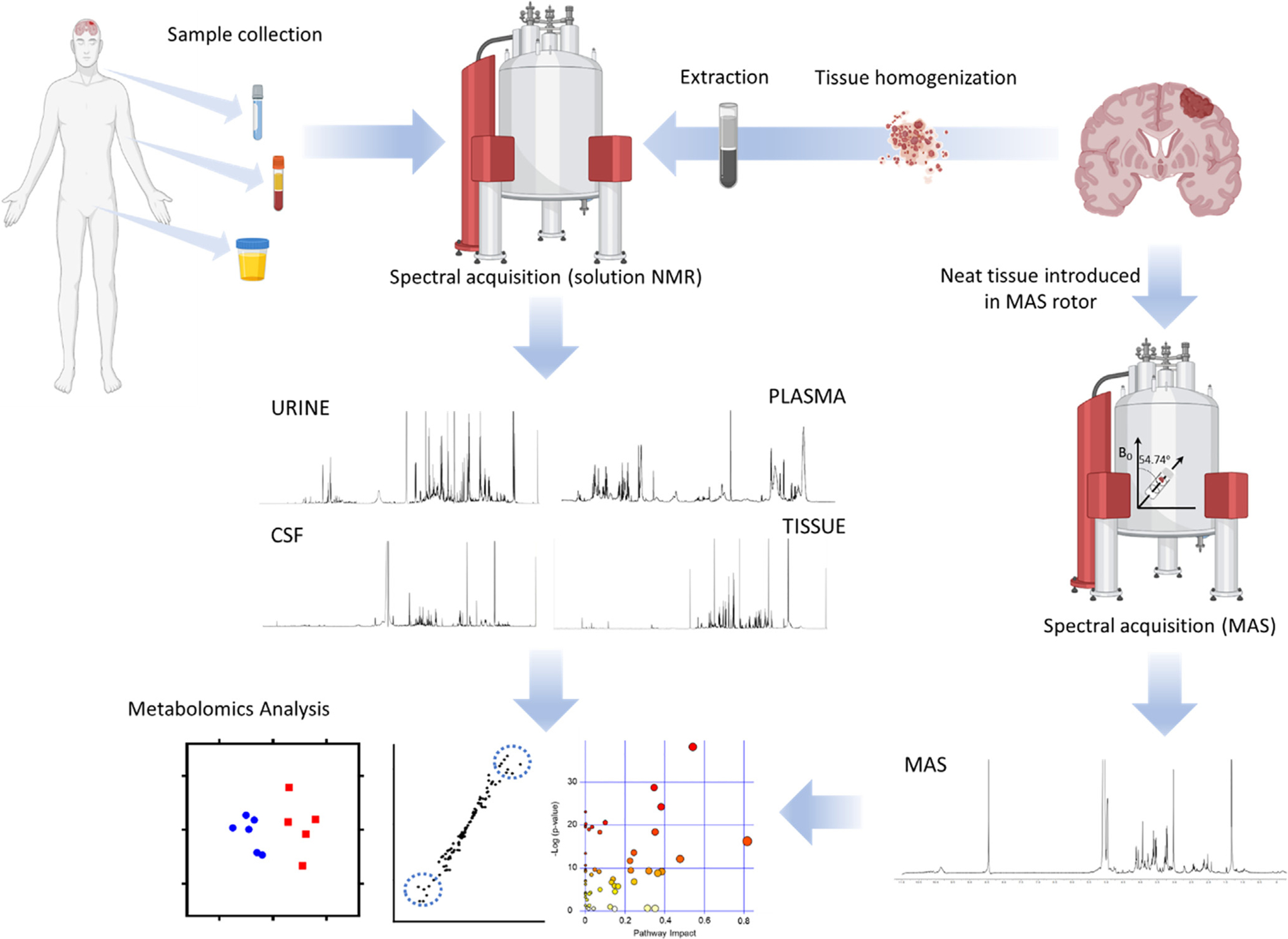
Workflow for metabolomics investigations of brain tumors. After sample (CSF, blood, urine) collection from patients or animal models the preparation steps are minimal, involving mixing the sample with D2O and a buffer, prior to transfer the sample to an NMR tube and the acquisition of the spectra. Analysis of tissue requires the mechanical homogenization of the specimen and subsequent extraction utilizing organic solvents to create a biphasic preparation after centrifugation. Both polar (upper phase) and organic (bottom phase) fractions are dried and reconstituted in a deuterated solvent. Alternatively, tissue can be directly inserted into the MAS rotor for spectral acquisition (MAS spectrum courtesy of Dr. Madhu Basetti, University of Cambridge). Spectra are then analyzed by quantifying the assigned metabolites or by bucketing of the spectra to generate datasets that can be employed for multivariate analysis. This approach usually involves the clustering of samples according to disease status or treatment based on their metabolic profiles, the search of dysregulated metabolites that can be utilized as biomarkers and the pathway analysis to assign the changes in metabolite levels to a specific metabolic route.
