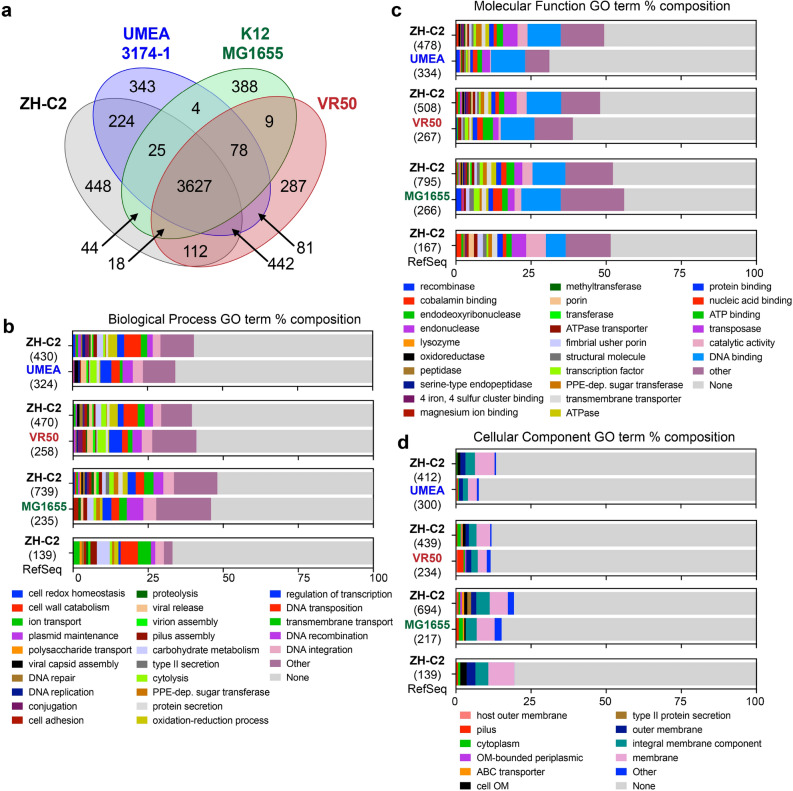
Figure 6.
Comparison of encoded E. coli ZH-C2 genes to reference non-pathogenic and pathogenic E. coli. Venn diagram of CD-HIT analysis depicting conservation and differences of gene products between E. coli strains ZH-C2, K-12 MG1655, UMEA 3174-1, and VR50 (a). Protein sequences were clustered at CD-HIT sequence similarity cut-off of 90%. Biological process (b), molecular function (c), and cellular component (d) GO Terms of unique genes (less than 70% coverage and/pr less than 90% identity) found in ZH-2 compared to MG1655, UMEA 3174-1, VR50, and the RefSeq reference genome collection. The number of genes used for each GO term analysis is in parentheses. GO Terms listed as “other” include < 2 genes and are expanded in the Supplementary Table 1.