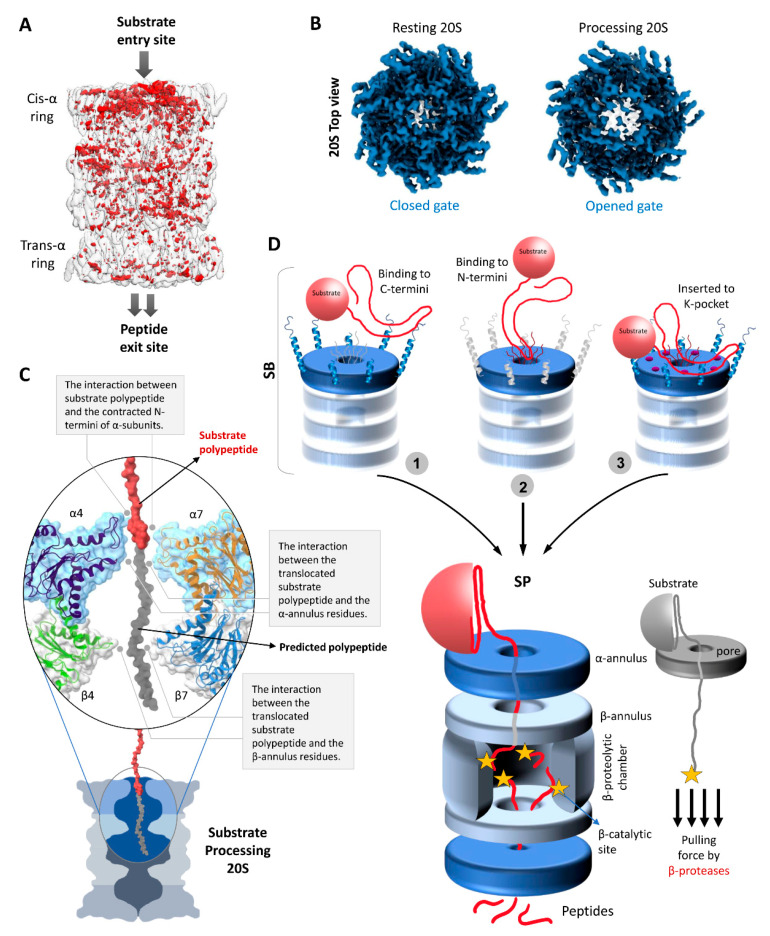
Figure 4.
Structural conformational changes during substrate processing and functional model of the 20S proteasome. (A) A differential map rendering the asymmetric pattern between the trans- and cis- α-rings of substrate-processing human 20S proteasomes [49]. (B) The top view electron density map exposing the cis α-ring of resting 20S proteasome vs. substrate processing 20S proteasome [49]. (C) Potential interactions of a substrate polypeptide with the contracted N-termini of α-subunits, α-annulus residues, and β-annulus residues during substrate processing state. The surface and embedded cartoon representation of α4/7- and β4/7 along with the substrate polypeptide (in red) image was generated using the model of substrate-bound human 26S proteasomes (PDB: 6msk). A putative extension of the substrate polypeptide is colored grey for illustration purposes. (D) A model mechanism of substrate processing by 20S proteasomes. In substrate accepting state, we propose that the unstructured portion of the substrate would recognize and interact either with (1) the protruding C-termini of α-subunits, or with (2) the long N-terminal loops of α-subunits, or (3) may insert into K-pockets of α-subunits. The pulling force of β-proteases aids unfolding of the remainder of the substrate against the α- and β-annulus (aperture) by a ratchet mechanism.