Abstract
Estimates show that 5–10% of breast cancer cases are hereditary, caused by genetic variants in autosomal dominant genes; of these, 16% are due to germline mutations in the BRCA1 and BRCA2 genes. The comprehension of the mutation profile of these genes in the Brazilian population, particularly in Amazonian Amerindian groups, is scarce. We investigated fifteen polymorphisms in the BRCA1 and BRCA2 genes in Amazonian Amerindians and compared the results with the findings of global populations publicly available in the 1000 Genomes Project database. Our study shows that three variants (rs11571769, rs144848, and rs11571707) of the BRCA2 gene, commonly associated with hereditary breast cancer, had a significantly higher allele frequency in the Amazonian Amerindian individuals in comparison with the African, American, European, and Asian groups analyzed. These data outline the singular genetic profiles of the indigenous population from the Brazilian Amazon region. The knowledge about BRCA1 and BRCA2 variants is critical to establish public policies for hereditary breast cancer screening in Amerindian groups and populations admixed with them, such as the Brazilian population.
Keywords: hereditary breast cancer, BRCA1, BRCA2, indigenous populations, Native Americans, Brazil
1. Introduction
Breast cancer (BC) is the most common type of cancer in women, excluding non-melanoma cancer. By 2040, more than three million people around the globe will be affected by this cancer type [1]. In Brazil, the National Cancer Institute (INCA) estimates new cases of breast cancer for each year in the 2020–2022 period, this being the most frequent cancer site—excluding non-melanoma cancer—in Brazilian females, including those from the northern region [2].
The BRCA1 and BRCA2 genes act as tumor suppressors by maintaining genomic stability and DNA repair [3]. Mutations in these genes are related to a higher susceptibility to BC, which can increase more than 80% of the chances of developing this cancer type during life [4].
About 5–10% of BC cases are due to hereditary factors, which are caused by variants in autosomal dominant genes; of these, 16% can be attributed to germline mutations in the BRCA1 and BRCA2 genes [5]. The frequency of allelic variants of these genes varies widely between different population groups. For example, in Ashkenazi Jews, approximately 1 in 40 individuals has one of three specific founder mutations (187delAG or 5385insC in BRCA1 or 6174delT in BRCA2) [6]. Despite the known population variability of variants, little is known about the profile of mutations in the BRCA1 and BRCA2 genes in the Brazilian population, particularly in Amazonian Amerindian groups.
According to the Brazilian Institute of Geography and Statistics (IBGE), indigenous people represent 0.47% of the Brazilian population, summing a total of 896,917 individuals [7]; nevertheless, reports about cancer prevalence in this population are still rare. In other countries, such as Australia and New Zealand, records indicate that cancer is the second leading cause of death among indigenous people [8]. There are no studies that assess the impact of genetic variants in Amazonian Amerindian populations and the risk of developing breast cancer.
This study characterizes the molecular profile of the BRCA1 and BRCA2 genes by analyzing the exome of Amerindian populations from the Brazilian Amazon. We aimed to describe variants that may affect the development of breast cancer in Amerindian populations and to compare these data with populations of distinct ancestry background.
2. Materials and Methods
2.1. Study and Reference Populations
The study population is composed of 64 Amerindians from the Amazon region of northern Brazil who represent 12 different Amazonian ethnic groups: five Asurini of the Xingu, seven Arara, six Araweté, 16 Asurini of the Tocantins, eight Awa-Guajá, two Kayapó/Xikrin, five Zo’é, ten Wajãpi, one Karipuna, one Phurere, one Munduruku and two Juruna. All individuals were grouped as the Indigenous group (IND). The Amerindian individuals share no family relationships and do not present breast cancer or cases of breast cancer in their families. The genetic ancestry was obtained through a panel of 64 informative ancestry markers (IAM), as described by Ramos et al. 2016 [9]. Additional information about this population can be found in [10].
All participants of the study and their ethnic group leaders signed a free-informed consent. The recruitment period for participants was from September 2017 to December 2018. The study was approved by the National Committee for Ethics in Research (CONEP) and the Research Ethics Committee of the UFPA Tropical Medicine Center, under CAAE number 20654313.6.0000.5172.
We compared our results with those of populations from other countries available at the phase 3 release of the 1000 Genomes Database (available at http://www.1000genomes.org).
These populations include 5203 individuals of African (AFR), 5789 of American (AMR), 4327 of East Asian (EAS), 33,370 of European (EUR), and 8256 of South Asian (SAS) descent. As indicated in [11], for the samples with ancestry from Europe, East Asia, and South Asia, populations across the geographic range had about 1% FST; the populations from Africa are related to the Yoruba and are therefore not comprehensive within Africa; for populations in the Americas, the samples are from two populations with primarily African and European ancestry (people with African Ancestry in southwest USA (ASW) and the African Caribbean in Barbados (ACB) and four populations (People with Mexican Ancestry in Los Angeles, CA, USA (MXL), Colombians in Medellin, Colombia (CLM), Puerto Ricans in Puerto Rico (PUR), Peruvians in Lima, Peru (PEL) with a wide range of European, African, and indigenous American ancestry chosen to represent the wide variation in ancestry proportions observed in North, Central, and South America.
2.2. Extraction of the DNA and Preparation of the Exome Library
The DNA extraction was performed by the phenol-chloroform method [12]. The quantification and integrity of genetic material were analyzed by a Nanodrop-8000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA) and electrophoresis in 2% agarose gel, respectively.
The exome libraries were prepared using the Nextera Rapid Capture Exome (Illumina®, San Diego, CA, USA) and SureSelect Human All Exon V6 (Agilent) kits. The sequencing reactions were run in the NextSeq 500® platform (Illumina®, San Diego, CA, USA) using the NextSeq 500 High-output v2 300 cycle kit (Illumina®, San Diego, CA, USA).
2.3. Bioinformatic Analysis
The bioinformatic analysis was performed as previously described in Rodrigues and colleagues [10].
2.4. Statistical Analyses
The allele frequencies of the IND populations were obtained by gene counting and compared with the other study populations (AFR, EUR, AMR, EAS, and SAS). Fisher’s exact test was used to test the difference in frequencies between the populations. A p-value ≤ 0.05 was considered significant. The interpopulation variability of the polymorphisms was assessed using Wright’s fixation index (FST). All analyses were run in RStudio v.3.5.1.
2.5. Selection of Variants
The selection of variants for subsequent analyses was based on two main criteria: (a) minimum of 10 reads of coverage (fastx_tools v.0.13-http://hannonlab.cshl.edu/fastx_toolkit/); (b) the variant impact should be either modifier, moderate or high, according to SNPeff classification (https://pcingola.github.io/SnpEff/). A total of 41 variants were found in the BRCA1 and BRCA2 genes, which are described in Supplementary Table S1. The analyses were directed to 15 variants that met all specifications of the selection criteria.
3. Results
Fifteen variants were identified, eight in the BRCA1 gene and seven in the BRCA2 gene, in the individuals analyzed (Table 1). This table contains the characteristics of these variants, including their reference number, chromosomal region, nucleotide exchange, impact predicted by the SNPeff software, and the allele frequency referring to the IND population and the five continental populations present in the 1000 Genomes platform (AFR, AMR, EAS, EUR, and SAS). Among the selected polymorphisms, five have predicted impact as modifier and ten as moderate.
Table 1.
Description of the variants in the BRCA1 and BRCA2 genes in the Amerindian individuals (IND) and continental populations (African (AFR), American population (AMR), East Asian (EAS), European (EUR), and South Asian (SAS)) described in the 1000 genomes database.
| Gene | SNP ID | Region | Change in Nucleotide | Impact Predicted by SNPeff | Minor Allele Frequencies | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| IND | AFR | AMR | EAS | EUR | SAS | |||||
| BRCA1 | rs799923 | Intronic | G > A | Modifier | 0.016 | 0.0389 | 0.0845 | 0.0007 | 0.2299 | 0.1797 |
| BRCA1 | rs16941 | CDS | T > C | Moderate | 0.453 | 0.1797 | 0.3083 | 0.3777 | 0.3254 | 0.4996 |
| BRCA1 | rs16942 | CDS | T > C | Moderate | 0.453 | 0.2358 | 0.3141 | 0.3827 | 0.3259 | 0.4999 |
| BRCA1 | rs1799966 | Other a | T > C | Moderate | 0.453 | 0.2397 | 0.3138 | 0.3805 | 0.3268 | 0.5001 |
| BRCA1 | rs12516 | 3UTR | G > A | Modifier | 0.24 | 0.3012 | 0.3433 | 0.4286 | 0.3841 | 0.4839 |
| BRCA1 | rs799905 | Intronic | G > C | Modifier | 0.4754 | 0.7789 | 0.4468 | 0.4403 | 0.3796 | 0.5116 |
| BRCA1 | rs1799949 | Other a | G > A | Moderate | 0.4531 | 0.2364 | 0.3138 | 0.3783 | 0.3252 | 0.5002 |
| BRCA1 | rs799917 | CDS | G > A | Moderate | 0.4841 | 0.8193 | 0.3435 | 0.378 | 0.3341 | 0.5285 |
| BRCA2 | rs11571769 b | CDS | G > A | Moderate | 0.1111 | 0.0012 | 0.0349 | 0 | 0.0044 | 0.014 |
| BRCA2 | rs144848 b | CDS | A > C | Moderate | 0.5078 | 0.1249 | 0.3049 | 0.2728 | 0.2818 | 0.3558 |
| BRCA2 | rs1799943 | 5UTR | G > A | Modifier | 0.0937 | 0.1005 | 0.1895 | 0.3805 | 0.2581 | 0.2829 |
| BRCA2 | rs11571651 | Intronic | G > T | Modifier | 0 | 0.0238 | 0.0633 | 0.0994 | 0.0335 | 0.1157 |
| BRCA2 | rs1799944 | CDS | A > G | Moderate | 0.1406 | 0.0427 | 0.0672 | 0.1002 | 0.035 | 0.1151 |
| BRCA2 | rs11571707 b | CDS | T > C | Moderate | 0.3203 | 0.0026 | 0.1409 | 0.0036 | 0.0003 | 0.0018 |
| BRCA2 | rs766173 | CDS | A > C | Moderate | 0.1379 | 0.024 | 0.0667 | 0.1002 | 0.035 | 0.1159 |
a NEXT_PROT (modified-residue:phosphoserine); b variants related to hereditary breast and ovarian cancer syndrome (HBOC) accordingly to ClinVar.
The multidimensional scale analysis (MDS) using the FST values (Supplementary Table S2) for each pairwise comparison for the 15 variants in the BRCA1 and BRCA2 genes revealed the existence of three major groups (Figure 1): the African population (AFR) is completely isolated, showing greater genetic diversity; the European (EUR), East Asian (EAS) and South Asian (SAS) populations have grouped in the top right corner; the third group was composed of the American population (AMR) and the Amerindians of this study (IND).
Figure 1.
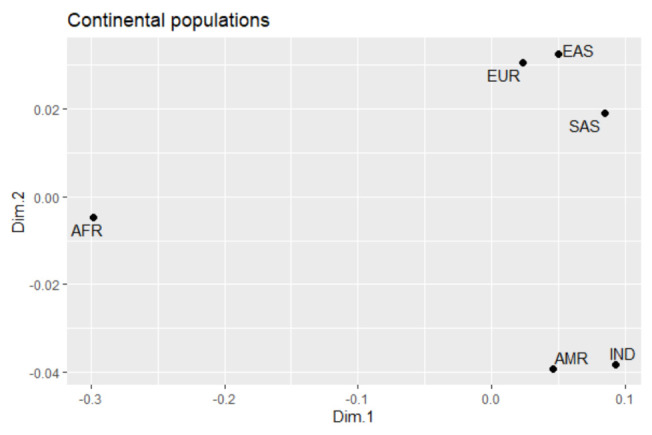
Multidimensional scaling plot illustrating the grouping of ethnic populations according to the genetic profile of the 15 variants in the BRCA1 and BRCA2 genes.
In addition, the frequencies of these 15 variants were compared with the global populations using Fisher’s exact test (Table 2). The molecular profile of the indigenous individuals of this study varies considerably from African, American, and European populations.
Table 2.
Comparison between the allelic frequency of the study population Indian (IND) and continental populations (African (AFR), American (AMR), East Asian (EAS), European (EUR), and South Asian (SAS)) described in the 1000 Genomes database.
| Gene | SNP Id | IND vs. AFR * | IND vs. AMR * | IND vs. EAS * | IND vs. EUR * | IND vs. SAS * |
|---|---|---|---|---|---|---|
| BRCA1 a | rs799923 | 0.5186 | 0.0409 | 0.05706 | 1.705 × 10−6 | 9.866 × 10−5 |
| BRCA1 | rs16941 | 5.371 × 10−7 | 0.01994 | 0.2428 | 0.03282 | 0.5307 |
| BRCA1 | rs16942 | 1.614 × 10−4 | 0.02113 | 0.2474 | 0.03306 | 0.5307 |
| BRCA1 | rs1799966 | 1.96 × 10−4 | 0.02108 | 0.2449 | 0.04436 | 0.5307 |
| BRCA1 | rs12516 | 0.663 | 0.3983 | 0.06793 | 0.1548 | 0.01586 |
| BRCA1 a | rs799905 | 4.308 × 10−7 | 0.6986 | 0.6052 | 0.1459 | 0.6085 |
| BRCA1 | rs1799949 | 1.657 × 10−4 | 0.02108 | 0.2433 | 0.03272 | 0.5307 |
| BRCA1 | rs799917 | 1.877 × 10−9 | 0.02391 | 0.0917 | 0.01618 | 0.5305 |
| BRCA2 | rs11571769 b | 4.044 × 10−11 | 0.00679 | 8.917 × 10−14 | 1.71 × 10−8 | 3.626 × 10−5 |
| BRCA2 b | rs144848 | 1.567 × 10−13 | 9.83 × 10−4 | 6.84 × 10−5 | 1.429 × 10−4 | 0.01323 |
| BRCA2 | rs1799943 | 1 | 0.05333 | 3.735 × 10−7 | 0.00149 | 3.912 × 10−4 |
| BRCA2 | rs11571651 | 1 | 0.3988 | 0.1629 | 1 | 0.1031 |
| BRCA2 | rs1799944 | 0.001716 | 0.03866 | 0.2925 | 3.891 × 10−4 | 0.5529 |
| BRCA2 a,b | rs11571707 | <2.2 × 10−16 | 0.0002095 | <2.2 × 10−16 | <2.2 × 10−16 | <2.2 × 10−16 |
| BRCA2 | rs766173 | 8.935 × 10−5 | 0.0563 | 0.3749 | 9.218 × 10−4 | 0.5394 |
a intronic variant; b variants related to hereditary breast and ovarian cancer syndrome (HBOC); * p-value obtained by Fisher’s exact test; bold: significant result (p-value ≤ 0.05).
The EUR population stands out as the one that most presented variants with significant differences (12 out of 15) to the Amerindian population; of which six were in the BRCA1 genes (rs799923, rs16941, rs16942, rs1799966, rs1799949, and rs799917) and six in the BRCA2 gene (rs11571769, rs144848, rs1799943, rs1799944, rs11571707, and rs766173).
Regarding the AFR population, six polymorphisms were found to be significantly divergent in the BRCA1 gene (rs16941, rs16942, rs1799966, rs799905, rs1799949, and rs799917) and five in the BRCA2 gene (rs11571769, rs144848, rs1799944, rs11571707, and rs766173), summing a total of 11 significantly different variants from the IND population.
The American population (AMR) presented 10 statistically different polymorphisms in relation to the IND population: six in the BRCA1 gene (rs799923, rs16941, rs16942, rs1799966, rs1799949 and rs799917) and four in the BRCA2 gene (rs11571769, rs144848, rs1799944 and rs11571).
Moreover, the comparison between IND and SAS and EAS populations showed the same number of significantly different SNPs (6 out of 15). Two variants in the BRCA1 gene (rs12516 and rs799917) and four in the BRCA2 gene (rs11571769, rs144848, rs1799943, and rs11571707) were divergent in the EAS population. The SAS population also showed two divergent polymorphisms in the BRCA1 gene (rs799923 and rs12516) and four in the BRCA2 gene (rs11571769, rs144848, rs1799943, and rs11571707).
It is worth mentioning that three variants in the BRCA2 gene (rs11571769, rs144848, and rs11571707) presented a unique frequency in the Amazonian Amerindian populations, diverging from that observed in all other evaluated populations. The distribution of the mutant alleles in the indigenous ethnicities can be consulted in Table 3. No mutant homozygotes were observed for rs11571769 in the IND group, whereas for the rs11571707 and the rs144848 variants, 8 and 16 individuals had homozygous mutant genotypes, respectively.
Table 3.
The distribution of mutant alleles among the indigenous groups.
| Amerindian Group | N. of Individuals | Number of Variant Alleles | ||
|---|---|---|---|---|
| rs11571769 | rs11571707 | rs144848 | ||
| Asurini of the Xingu | 5 | 2 | 4 | 4 |
| Arara | 7 | 0 | 8 | 8 |
| Araweté | 6 | 4 | 4 | 4 |
| Asurini of the Tocantins | 16 | 0 | 5 | 23 |
| Awa-Guajá | 8 | 1 | 4 | 6 |
| Kayapó/Xikrin | 2 | 1 | 1 | 1 |
| Zo’é | 5 | 4 | 6 | 6 |
| Wajãpi | 10 | 1 | 8 | 10 |
| Karipuna | 1 | 0 | 0 | 0 |
| Phurere | 1 | 0 | 1 | 1 |
| Munduruku | 1 | 1 | 1 | 1 |
| Juruna | 2 | 0 | 0 | 0 |
The allele frequency of these polymorphisms is substantially higher in indigenous individuals when compared to individuals of other ancestries. The rs11571707 (NM_000059.4:c.7469T>C; NP_000050.3:p.Ile2490Thr) showed the greatest allele frequency in the study subjects (32.03%-41 mutant alleles) in contrast to the world populations (except the AMR) where the frequencies are less than 1%. Similarly, the rs144848 (NM_000059.4:c.1114A>C, NP_000050.3:p.Asn372His) also had a significantly high-frequency in indigenous people (50.78%-65 mutant alleles), whereas the other populations showed a mean frequency lower than 27%. Additionally, the rs11571769 (NM_000059.4:c.8851G>A NP_000050.3:p.Ala2951Thr) showed a frequency of 11.1% (14 mutant alleles) in the IND individuals in contrast to a mean frequency of 1% in the other populations.
4. Discussion
Recently, in Latin America and Brazil, cancer has been growing gradually among the indigenous people [8,13,14]. Around 10% of indigenous deaths in Latin America are caused by some neoplasm, of which breast cancer accounts as the main responsible [15,16].
In 2003/2005, the National Health Foundation (FUNASA) reported the diagnosis of 45 cases of breast cancer in indigenous people in Brazil [17]. In addition, according to Freitas et al. (2015) [12], in 2000 and 2010, there were fifteen cases of death from breast cancer in Native Americans (13.31% in 2000 and 5.01% in 2010). There is a lack of evidence in the literature about current information concerning breast cancer in the population of the Americas, probably due to an underreporting of cases in local communities [8,14]. Furthermore, the traditional populations of Latin America constitute a complex study group due to their human history of miscegenation, which confers them with high levels of interpopulation genetic diversity [10].
The detection of germline mutations is essential for clinical control and early screening for breast cancer. Nevertheless, the accumulated knowledge about mutation profiles in the BRCA1 and BRCA2 genes in the Brazilian population, particularly in American Amazonian populations, is insufficient [18]. The study population of the Online Archive of Brazilian Mutations (ABraOM) database, which is composed of Brazilian individuals from the southeast of the country, who form a very heterogeneous population with higher degrees of European ancestry, followed by African ancestry and only 10% of Amerindian ancestry [9], does not represent a good resource for the care of individuals from the isolated and homogeneous Amerindian groups from the Brazilian Amazon.
Previous studies report a significant heterogeneity in the mutation burden of the BRCA1 and BRCA2 genes between different countries and even among regions of the same country (for example, in Brazil), which impairs the effectiveness and applicability of the use of already existing panels in Amerindian and Brazilian populations [18,19].
Therefore, our study aimed to characterize the molecular profile of the BRCA1 and BRCA2 genes through the analysis of the exome of Amerindian populations from the Brazilian Amazon region, to describe variants that can be associate with the risk of breast cancer, and to compare the results with publicly available data about global populations.
This is the first study to investigate the BRCA1 and BRCA2 genes in Amazonian Amerindians, a genetically distinct population that is unrepresented in genomic investigations. The Amerindian ancestry contribution in Brazil has an average of 17%, while in the Amazon region, this level of admixture is approximately 30%, standing out as the region with the greatest Amerindian genetic contribution of Brazil [20,21].
Of the 41 variants found in the exome analysis of the BRCA1 and BRCA2 genes, our study analyzed 15 that could be potentially related to the risk of developing breast cancer. Among the variants investigated, ten of them show a moderate impact, of which nine are classified as missense and one as synonymous; the five remaining variants had a modifier impact, being three intronic, one in the 3′-UTR region and one in the 5′-UTR region.
Among the eight single nucleotide polymorphisms (SNP) of the BRCA1 gene, the rs799923, rs1799966 and rs799917 have been associated with the increased risk of BC in Asians [22,23]. In contrast, the rs16942 and rs1799949 variants have been associated with the predisposition to BC in the African population [24,25]. Other investigations linked the rs16941 polymorphism with susceptibility of BC in Caucasians [24]. The rs799917 and rs12516 polymorphisms in the BRCA1 gene have been little explored in genetic association studies [26,27].
Regarding the seven SNPs of the BRCA2 gene, there is an association of the development of BC with the rs115771651, rs766173, and rs1799943 variants in the Asian population [28,29] and rs1799944 and rs1799943 polymorphism in Caucasians [30,31]. The other three polymorphisms (rs11571707, rs11571769, rs144848) of the BRCA2 gene evaluated here are related to hereditary breast cancer [32,33]. All these three variants had high allelic frequencies in the Amerindian populations in comparison to the global populations analyzed.
The rs11571707 showed the greatest difference between the allele frequency of the IND individuals (32.03%) and the majority of the global populations, which had a frequency of less than 1%, except the AMR that showed a frequency of 14%. However, the AMR population of the 1000 Genomes Project is composed mainly of Latinos, who present levels of Amerindian miscegenation [11]. According to Solano et al. (2012), the rs11571707 variant is often associated with hereditary breast and ovarian cancer syndrome (HBOC) in Latin Americans [34,35]. As observed in Table 3, the rs11571707 showed 12.5% (8 out of 64) IND individuals with homozygous mutant genotype.
Similarly, the rs144848 had a significantly higher frequency in the Amazonian Amerindian peoples (50.78%) when compared to Americans (30.5%), Europeans (28.2%), South Asians (35.5%), East Asians (27.3%), and, particularly, Africans (12%). This variant has been pointed to act as a factor that confers a moderate risk for the development of BC [36,37,38]. Studies have already observed a relationship between the mutant homozygous genotype and the increased risk of breast cancer in women under 60 years of age [36] and family members of breast cancer patients [39]. As shown in Table 3, 25% (16 out of 64) IND individuals were homozygous mutant for the rs144848 variant.
Furthermore, the rs11571769 showed a frequency of 11.1% in the IND group of this study in contrast to a frequency of 3% in AMR, 1.4% in SAS, less than 1% in both AFR and EUR (the variant allele was not reported in the EAS populations). According to a study conducted with patients with HBOC patients in Brazil, the rs11571769 was identified as a predictor of pathogenicity for BC development [40,41]. Our analyses did not report any homozygous mutant individual for the rs11571769 variant in the IND group.
The Amerindians from the Amazon region have a unique genetic profile because of stochastic processes resulting from a long process of geographic isolation and inbreeding [42]. Additionally, some small groups migrated from settled areas to uninhabited territories, giving rise to the first indigenous communities [36]. The genetic drift effect in these populations was even more driven by the small population size and inbreeding relationships, resulting in higher numbers of deleterious homozygous genotypes in the Native American populations [37,38].
The MDS analysis resulted in the formation of a single group formed by AMR and IND, which was caused by their genetic similarity (Fst = 0.03669). The population of Americans is known for their trichotomous genetic admixture with ancestral contributions of European colonists, African immigrants, and Native Amerindians [43,44]. Thus, since the Amerindians were one of the forming group of the Latin Americans, they have similarities in their genomic profile, including the BRCA1 and BRCA2 genes, a fact that can be extremely relevant in the likely high frequency of variants linked to breast cancer in these populations [45,46].
In contrast, the comparative analysis of the BRCA1 and BRCA2 variants in the AMR population showed 10 polymorphisms with significant statistical differences to the IND population. The sample of the American population of the 1000 Genomes Project includes several countries of Latin America, such as Mexico, Peru, Colombia, and Puerto Rico [11]. Although there is a degree of genetic similarity between AMR and IND populations, the genomic profiles of these countries indicate considerable heterogeneity in terms of Amerindian ancestry contribution, resulting from the different historical aspects of their formation and their degree of genetic admixture, which may explain the differences found in the BRCA1 and BRCA2 gene variants [10,47,48,49].
5. Conclusions
The exome analysis of the BRCA1 and BRCA2 genes in the Amazonian Amerindian populations reported 15 polymorphisms potentially capable of increasing susceptibility to breast cancer. Among them, we highlight the rs11571707, rs144848, and rs11571769 variants of the BRCA2 gene due to their association with the hereditary breast cancer onset. This study may help the establishment of future public policies for early breast cancer screening in Amerindian populations and also Brazilian individuals with high levels of Amerindian ancestry contributions.
Acknowledgments
We thank Núcleo de Pesquisas em Oncologia- NPO and Laboratório de Genética Humana e Médica- LGHM for technical support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/2/142/s1, Table S1: Description of all variants in the BRCA1 and BRCA2 genes found in the Amerindian individuals (IND), Table S2: Pairwise FST among Amerindians (IND) and the five continental populations from the 1000 Genomes database.
Author Contributions
Conceptualization, E.A.F.D. and J.A.G.M.; methodology, J.C.G.R.; software: J.E.K. and S.J.d.S.; formal analysis, J.A.G.M. and J.C.G.R.; investigation, E.A.F.D., J.A.G.M. and M.S.C.R.C.; resources, Â.R.-d.-S. and J.F.G.; writing—original draft preparation, E.A.F.D. and J.A.G.M.; writing—review and editing, E.A.F.D., J.A.G.M., J.C.G.R. and N.P.C.d.S.; supervision, N.P.C.d.S., M.R.F., P.P.d.A., S.E.B.d.S. and R.M.R.B.; project administration, J.F.G., S.E.B.d.S. and N.P.C.d.S.; funding acquisition, J.F.G., Â.R.-d.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (http://www.cnpq.br); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (https://www.gov.br/capes/pt-br); Pró-Reitoria de Pesquisa e Pós-Graduação da UFPA—PROPESP (http://www.propesp.ufpa.br) and Fundação Amazônia de Amparo a Estudos e Pesquisas- FAPESPA (http://www.fapespa.pa.gov.br). This work is part of Rede de Pesquisa em Genômica Populacional Humana (Biocomputacional-Protocol no. 3381/2013/CAPES).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by Ethics Committee of the by the National Committee for Ethics in Research (CONEP) and the Research Ethics Committee of the UFPA Tropical Medicine Center (CAAE number: 20654313.6.0000.5172).
Informed Consent Statement
All participants of the study and their ethnic group leaders signed a free-informed consent.
Data Availability Statement
The dataset used in this study is publicly available. The name of the repository and accession number(s) can be found at https://doi.org/10.6084/m9.figshare.13623989.v1.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Instituto Nacional do Câncer José Alencar Gomes da Silva (INCA) Estimativa 2020: Incidência de Câncer no Brasil. Ministério da Saúde; Brasilia, Brasil: 2019. [Google Scholar]
- 3.Wendt C., Margolin S. Identifying breast cancer susceptibility genes—A review of the genetic background in familial breast cancer. Acta Oncol. 2019;58:135–146. doi: 10.1080/0284186X.2018.1529428. [DOI] [PubMed] [Google Scholar]
- 4.Apostolou P., Fostira F. Hereditary breast cancer: The era of new susceptibility genes. BioMed Res. Int. 2013 doi: 10.1155/2013/747318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheikh A., Hussain S.A., Ghori Q., Naeem N., Fazil A., Giri S., Sathian B., Mainali P., Al Tamimi D.M. The spectrum of genetic mutations in breast cancer. Asian Pac. J. Cancer Prev. 2015;16:2177–2185. doi: 10.7314/APJCP.2015.16.6.2177. [DOI] [PubMed] [Google Scholar]
- 6.Rich T.A., Woodson A.H., Litton J., Arun B. Hereditary breast cancer syndromes and genetic testing. J. Surg. Oncol. 2015;111:66–80. doi: 10.1002/jso.23791. [DOI] [PubMed] [Google Scholar]
- 7.O Brasil Indígena. [(accessed on 9 April 2019)]; Available online: https://indigenas.ibge.gov.br/images/pdf/indigenas/folder_indigenas_web.pdf.
- 8.Moore S.P., Forman D., Piñeros M., Fernández S.M., de Oliveira Santos M., Bray F. Cancer in indigenous people in Latin America and the Caribbean: A review. Cancer Med. 2014;3:70–80. doi: 10.1002/cam4.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramos A.B.R., Mendes N.D., Tanikawa A.A., Amador M.A.T., dos Santos N.P.C., dos Santos S.E.B., Castelli E.C., Witkinda S.S., Silva M.G. Ancestry informative markers and selected single nucleotide polymorphisms in immunoregulatory genes on preterm labor and preterm premature rupture of membranes: A case control study. BMC Pregnancy Childbirth. 2016;16:30. doi: 10.1186/s12884-016-0823-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues J.C.G., Souza T.P.D., Pastana L.F., Ribeiro dos Santos A.M., Fernandes M.R., Pinto P., .Magalhães L. Identification of NUDT15 gene variants in Amazonian Amerindians and admixed individuals from northern Brazil. PLoS ONE. 2020;15:15. doi: 10.1371/journal.pone.0231651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1989. [Google Scholar]
- 13.Freitas Junior R.D., Soares L.R., Gonzaga C.M.R., Sousa A.L.L., Lima M.G.D., Branquinho L.W., Souza M.R.D. Mortalidade por câncer de mama em mulheres indígenas brasileiras. Rev. Bras. Mastol. 2015;5:41–45. doi: 10.5327/Z201500020002RBM. [DOI] [Google Scholar]
- 14.Montenegro R.A., Stephens C. Indigenous health in Latin America and the Caribbean. Lancet. 2006;367:1859–1869. doi: 10.1016/S0140-6736(06)68808-9. [DOI] [PubMed] [Google Scholar]
- 15.Hall G., Patrinos H.A. Indigenous Peoples, Poverty and Human Development in Latin America: 1994–2004. The World Bank; Washington, DC, USA: 2004. [Google Scholar]
- 16.Do Nascimento E.R., Wanderley A.V., Chalu-Pacheco F. Perfil clínico e epidemiológico do câncer entre os índios do estado do Pará, Brasil. Rev. Bras. Oncol. Clín. 2015;11:39. [Google Scholar]
- 17.Fundação Nacional de Saúde (FUNASA) Relatório de Gestão da FUNASA 2003/2005. Ministério da Saúde; Brasilia, Brasil: 2005. [Google Scholar]
- 18.Palmero E.I., Carraro D.M., Alemar B., Moreira M.A.M., Ribeiro-dos-Santos Â., Abe-Sandes K., Galvão H.C.R., Reis R.M., de Pádua Souza C., Campacci N., et al. The germline mutational landscape of BRCA1 and BRCA 2 in Brazil. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-27315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamps R., Brandão R.D., Bosch B.J., Paulussen A.D., Xanthoulea S., Blok M.J., Romano A. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int. J. Mol. Sci. 2017;18:308. doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos N.P., Ribeiro-Rodrigues E.M., Ribeiro-dos-Santos Â.K., Pereira R., Gusmão L., Amorim A., Santos S.E. Assessing individual interethnic admixture and population substructure using a 48–insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum. Mutat. 2010;31:184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- 21.Rodrigues de Moura R., Coelho A.V.C., de Queiroz Balbino V., Crovella S., Brandão L.A.C. Meta-analysis of Brazilian genetic admixture and comparison with other Latin America countries. Am. J. Hum. Biol. 2015;27:674–680. doi: 10.1002/ajhb.22714. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez A., Schoenfeld J.D., Nguyen P.L., Fiorentino M., Chowdhury D., Stampfer M.J., Sesso H.D., Giovannucci E., Mucci L.A., Shui I.M. Common variation in BRCA1 may have a role in progression to lethal prostate cancer after radiation treatment. Prostate Cancer Prostatic Dis. 2016;19:197–201. doi: 10.1038/pcan.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han M.R., Zheng W., Cai Q., Gao Y.T., Zheng Y., Bolla M.K., Michailidou K., Dennis J., Wang Q., Dunning A.M., et al. Evaluating genetic variants associated with breast cancer risk in high and moderate-penetrance genes in Asians. Carcinogenesis. 2017;38:511–518. doi: 10.1093/carcin/bgx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu G.P., Zhao Q., Wang D., Xie W.Y., Zhang L.J., Zhou H., Chen S.Z., Wu L.F. The association between BRCA1 gene polymorphism and cancer risk: A meta-analysis. Oncotarget. 2018;9:8681. doi: 10.18632/oncotarget.24064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagna T., Bonora E., Ouedraogo M.N.L., Fusco D., Zoure A.A., Bisseye C., Djigma F., Kafando J., Zongo N., Douamba Z., et al. Identification of BRCA1/2 p. Ser1613Gly, p. Pro871Leu, p. Lys1183Arg, p. Glu1038Gly, p. Ser1140Gly, p. Ala2466Val, p. His2440Arg variants in women under 45 years old with breast nodules suspected of having breast cancer in Burkina Faso. Biomol. Concepts. 2019;10:120–127. doi: 10.1515/bmc-2019-0015. [DOI] [PubMed] [Google Scholar]
- 26.Cox D.G., Simard J., Sinnett D., Hamdi Y., Soucy P., Ouimet M., Barjhoux L., Verny-Pierre C., McGuffog L., Healey S., et al. Common variants of the BRCA1 wild-type allele modify the risk of breast cancer in BRCA1 mutation carriers. Hum. Mol. Genet. 2011;20:4732–4747. doi: 10.1093/hmg/ddr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricks-Santi L., McDonald J.T., Gold B., Dean M., Thompson N., Abbas Wilson B., Kanaan Y., Naab T.J., Dunston G. Next-generation sequencing reveals high prevalence of BRCA1 and BRCA2 variants of unknown significance in early-onset breast cancer in African American women. Ethn. Dis. 2017;27:169. doi: 10.18865/ed.27.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sánchez-Chaparro M.M., Garza-Veloz I., Zayas-Villanueva O.A., Martinez-Fierro M.L., Delgado-Enciso I., Gomez-Govea M.A., Martínez-de-Villarreal L.E., Reséndez-Pérez D., Rodríguez-Sánchez I.P. Genetic Variants in the 3′UTR of BRCA1 and BRCA2 Genes and their Putative Effects on the microRNA Mechanism in Hereditary Breast and Ovarian Cancer. Diagnostics. 2020;10:298. doi: 10.3390/diagnostics10050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang F., Chen F., Xu J., Guan X. Identification and frequency of the rs12516 and rs8176318 BRCA1 gene polymorphisms among different populations. Oncol. Lett. 2016;11:2481–2486. doi: 10.3892/ol.2016.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayub S.G., Rasool S., Ayub T., Khan S.N., Wani K.A., Andrabi K.I. Mutational analysis of the BRCA2 gene in breast carcinoma patients of Kashmiri descent. Mol. Med. Rep. 2014;9:749–753. doi: 10.3892/mmr.2013.1862. [DOI] [PubMed] [Google Scholar]
- 31.De Silva S., Tennekoon K.H., Dissanayake A., De Silva K., Jayasekara L. Novel and reported pathogenic variants in exon 11 of BRCA2 gene in a cohort of Sri Lankan young breast cancer patients. Fam. Cancer. 2017;16:329–338. doi: 10.1007/s10689-016-9962-9. [DOI] [PubMed] [Google Scholar]
- 32.AL-Eitan L.N., Rababa’h D.M., Alghamdi M.A., Khasawneh R.H. Correlation between candidate single nucleotide variants and several Clinicopathological risk factors related to breast Cancer in Jordanian women: A genotype-phenotype study. J. Cancer. 2019;10:4647. doi: 10.7150/jca.33857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sapkota Y., Mackey J.R., Lai R., Franco-Villalobos C., Lupichuk S., Robson P.J., Kopciuk K., Cass C.E., Yasui Y., Damaraju S. Assessing SNP-SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0064896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loizidou M.A., Michael T., Neuhausen S.L., Newbold R.F., Marcou Y., Kakouri E., Daniel M., Papadopoulos P., Malas S., Hadjisavvas A., et al. DNA-repair genetic polymorphisms and risk of breast cancer in Cyprus. Breast Cancer Res. Treat. 2009;115:623–627. doi: 10.1007/s10549-008-0084-4. [DOI] [PubMed] [Google Scholar]
- 35.Trujillano D., Weiss M.E., Schneider J., Köster J., Papachristos E.B., Saviouk V., Zakharkina T., Nahavandi N., Kovacevic L., Rolfs A. Next-generation sequencing of the BRCA1 and BRCA2 genes for the genetic diagnostics of hereditary breast and/or ovarian cancer. J. Mol. Diagn. 2015;17:162–170. doi: 10.1016/j.jmoldx.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Spurdle A.B., Hopper J.L., Chen X., Dite G.S., Cui J., McCredie M.R., Giles G.G., Ellis-Steinborner S., Venter D.J., Newman B., et al. The BRCA2 372 HH genotype is associated with risk of breast cancer in Australian women under age 60 years. Cancer Epidemiol. Prev. Biomark. 2002;11:413–416. [PubMed] [Google Scholar]
- 37.Cifuentes-C L., Rivera-Herrera A.L., Barreto G. BRCA1 and BRCA2 mutations in a sample of breast and ovarian cancer families from the Colombian pacific. Colombia Méd. 2019;50:163–175. doi: 10.25100/cm.v50i3.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Da Costa E Silva Carvalho S., Cury N.M., Brotto D.B., de Araujo L.F., Rosa R.C.A., Texeira L.A., Plaça J.R., Marques A.A., Peronni K.C., Ruy P.d.C. Germline variants in DNA repair genes associated with hereditary breast and ovarian cancer syndrome: Analysis of a 21 gene panel in the Brazilian population. BMC Med. Genom. 2020;13:21. doi: 10.1186/s12920-019-0652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q., Guan R., Qiao Y., Liu C., He N., Zhang X., Zhang X., Jia X., Sun H., Yu J., et al. Association between the BRCA2 rs144848 polymorphism and cancer susceptibility: A meta-analysis. Oncotarget. 2017;8:39818. doi: 10.18632/oncotarget.16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasan T.N., Shafi G., Syed N.A., Alsaif M.A., Alsaif A.A., Alshatwi A.A. Lack of Association of BRCA1 and BRCA2 Variants with Breast Cancer in an Ethnic Population of Saudi Arabia, an Emerging High-Risk Area. Asian Pac. J. Cancer Prev. 2013;14:5671–5674. doi: 10.7314/APJCP.2013.14.10.5671. [DOI] [PubMed] [Google Scholar]
- 41.Solano A.R., Aceto G.M., Delettieres D., Veschi S., Neuman M.I., Alonso E., Chialina S., Chacón R.D., Mariani-Costantini R., Podestá E.J. BRCA1 and BRCA2 analysis of Argentinean breast/ovarian cancer patients selected for age and family history highlights a role for novel mutations of putative south-American origin. Springerplus. 2012;1:20. doi: 10.1186/2193-1801-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbina-Jara L.K., Rojas-Martinez A., Martinez-Ledesma E., Aguilar D., Villarreal-Garza C., Ortiz-Lopez R. Landscape of germline mutations in DNA repair genes for breast cancer in Latin America: Opportunities for PARP-like inhibitors and immunotherapy. Genes. 2019;10:786. doi: 10.3390/genes10100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva F.C., Lisboa B.C., Figueiredo M.C., Torrezan G.T., Santos É.M., Krepischi A.C., Rossi B.M., Achatz M.I., Carraro D.M. Hereditary breast and ovarian cancer: Assessment of point mutations and copy number variations in Brazilian patients. BMC Med. Genet. 2014;15:55. doi: 10.1186/1471-2350-15-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodrigues J.C.G., Fernandes M.R., Guerreiro J.F., da Silva A.L.D.C., Ribeiro-dos-Santos Â., Santos S., dos Santos N.P.C. Polymorphisms of ADME-related genes and their implications for drug safety and efficacy in Amazonian Amerindians. Sci. Rep. 2019;9:1–8. doi: 10.1038/s41598-019-43610-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mooney J.A., Huber C.D., Service S., Sul J.H., Marsden C.D., Zhang Z., Sabatti C., Ruiz-Linares A., Bedoya G. Understanding the hidden complexity of Latin American population isolates. Am. J. Hum. Genet. 2018;103:707–726. doi: 10.1016/j.ajhg.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S., Lewis Jr C.M., Jakobsson M., Ramachandran S., Ray N., Bedoya G., Rojas W., Parra M.V., Molina J.A., Gallo C., et al. Genetic variation and population structure in Native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandes G.C., Michelli R.A., Galvão H.C., Paula A.E., Pereira R., Andrade C.E., Felicio P.S., Souza C.P., Mendes D.R.P., Volc S., et al. Prevalence of BRCA1/BRCA2 mutations in a Brazilian population sample at-risk for hereditary breast cancer and characterization of its genetic ancestry. Oncotarget. 2016;7:80465. doi: 10.18632/oncotarget.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reich D., Patterson N., Campbell D., Tandon A., Mazieres S., Ray N., Parra M.V., Rojas W., Duque C., Mesa N., et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gravel S., Zakharia F., Moreno-Estrada A., Byrnes J.K., Muzzio M., Rodriguez-Flores J.L., Kenny E.E., Gignoux C.R., Maples B.K., Guiblet W., et al. Reconstructing Native American migrations from whole-genome and whole-exome data. PLoS Genet. 2013;9:e1004023. doi: 10.1371/journal.pgen.1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset used in this study is publicly available. The name of the repository and accession number(s) can be found at https://doi.org/10.6084/m9.figshare.13623989.v1.


