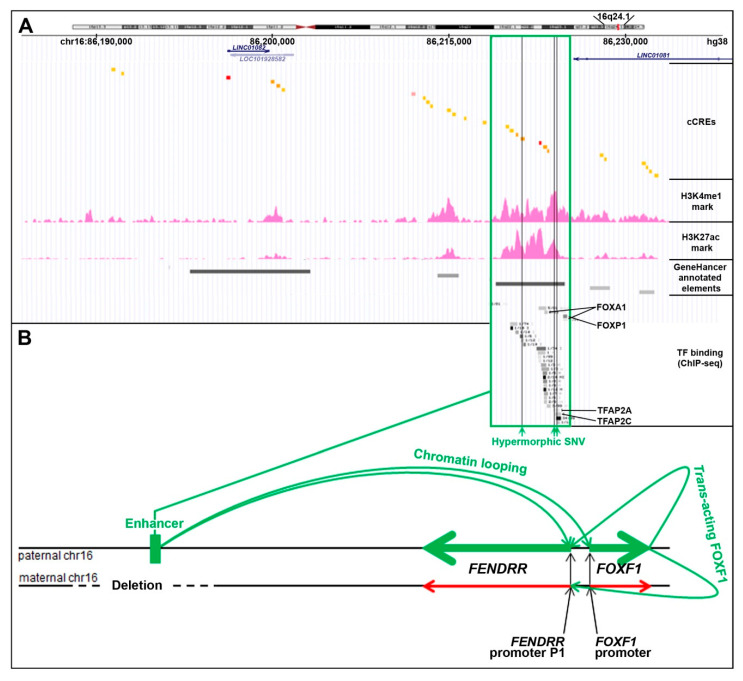
Figure 2.
Cis- and trans-regulation of the FENDRR expression in the lungs (modified from [24]). (A) FENDRR-FOXF1 distant enhancer region, located ~250 kb centromerically to the 3′ end of FENDRR (UCSC genome browser screenshot). The most essential part of this enhancer is shown in green frame. The enhancer features histone 3 modifications, usually found in active enhancers, and ChIP-seq-determined in HLFs binding sites for numerous transcription factors (TF): FOXA1, FOXP1, CEBPB, MAFK, RAD21, SMARCC1, CTCF, GTF2F1, KAP1, TBP, JUNs, EP300, STAT3, FOS, TFAP2A, and TFAP2C (ENCODE). Single nucleotide variants (SNVs, vertical green lines) that map to the essential part of the enhancer have been proposed to increase activity of the undeleted allele of the enhancer and mitigate ACDMPV phenotype in patients with heterozygous CNV deletions of the enhancer. (B) Scheme of mono-allelic expression of FENDRR from the paternally inherited chromosome 16 in the presence of a heterozygous CNV deletion of the maternal allele of the FENDRR-FOXF1 enhancer. The drawing shown is not to scale.