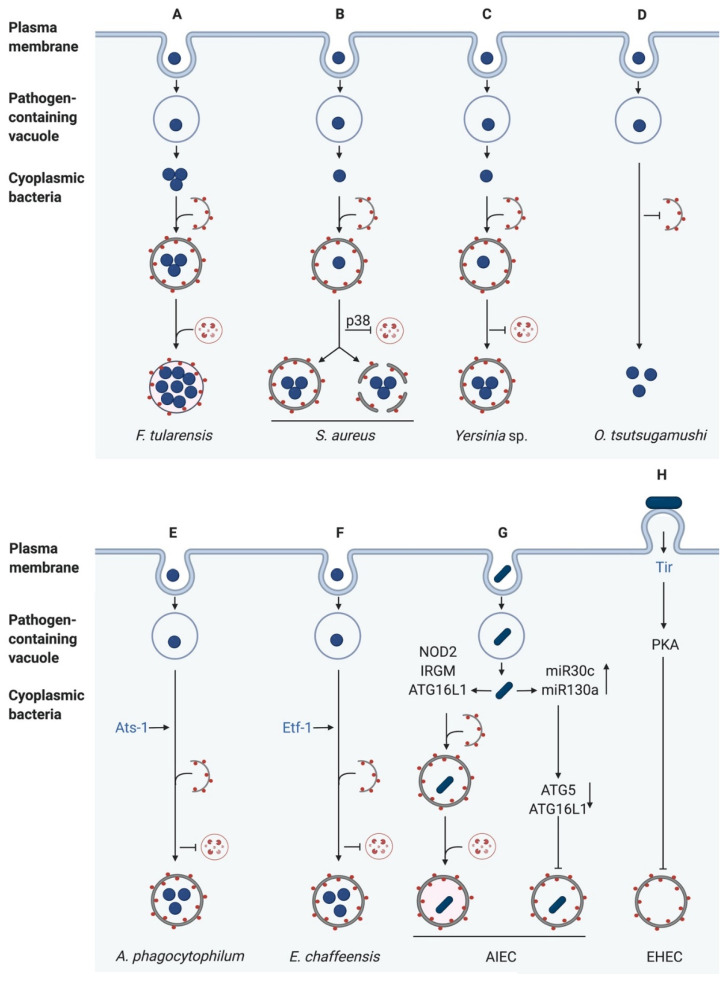
Figure 4.
Bacterial evasion or subversion of autophagy pathways. (A) Francisella tularensis escapes from the phagosome to replicate in the host cytoplasm. Afterwards, it enters the autophagy pathway and replicates in autophagosomes. (B) Staphylococcus aureus inhibits the autophagosome lysosome fusion through p38 MAPK phosphorylation. Then, it either replicates in autophagosomes or escapes from the autophagosome in an unknown mechanism. (C) Yersinia sp. block the fusion with lysosomes. Thus, Yersinia creates a replicative niche in autophagosomes. (D) After infection of host cells, O. tsutsugamushi induces autophagy, but the bacteria can prevent their uptake into the autophagosome. (E) Anaplasma phagocytophilum secretes Anaplasma translocated substrate-1 (Ats-1) to induce autophagy. However, the bacteria-containing inclusions fuse with autophagosomes to form amphisomes, delivering nutrients for bacterial replication. (F) Ehrlichia chaffeensis translocates Ehrlichia translocated factor-1 (Etf-1) to induce Rab5-regulated autophagy. This leads to the generation of amphisomes to provide nutrients for bacterial growth. (G) While intestinal epithelial cells induce autophagy after invasion of Adherent-invasive E. coli (AIEC) due to NOD2 activation, AIEC upregulates host microRNAs to subvert autophagy. (H) After Adhesion, Enterohaemorrhagic E. coli (EHEC) expresses its Type III secretion system (T3SS), leading to the translocation of translocated intimin receptor (Tir), which activates Protein kinase A (PKA) and thus blocks autophagy formation. LC3-II is displayed as red dots, and bacterial effector proteins are in blue (created with biorender.com).