Abstract
Background
Gastric cancer (GC) is considered one of the most lethal malignancies worldwide, which is accompanied by a poor prognosis. Although reports regarding the importance of cancer stem cell (CSC) markers in gastric cancer progression have rapidly developed over the last few decades, their clinicopathological and prognostic values in gastric cancer still remain inconclusive. Therefore, the current meta-analysis aimed to quantitatively re-evaluate the association of CSC markers expression, overall and individually, with GC patients’ clinical and survival outcomes.
Methods
Literature databases including PubMed, Scopus, ISI Web of Science, and Embase were searched to identify the eligible articles. Hazard ratios (HRs) or odds ratios (ORs) with 95% confidence intervals (CIs) were recorded or calculated to determine the relationships between CSC markers expression positivity and overall survival (OS), disease-free survival (DFS)/relapse-free survival (RFS), disease-specific survival (DSS)/ cancer-specific survival (CSS), and clinicopathological features.
Results
We initially retrieved 4,425 articles, of which a total of 66 articles with 89 studies were considered as eligible for this meta-analysis, comprising of 11,274 GC patients. Overall data analyses indicated that the overexpression of CSC markers is associated with TNM stage (OR = 2.19, 95% CI 1.84–2.61, P = 0.013), lymph node metastasis (OR = 1.76, 95% CI 1.54–2.02, P < 0.001), worse OS (HR = 1.65, 95% CI 1.54–1.77, P < 0.001), poor CSS/DSS (HR = 1.69, 95% CI 1.33–2.15, P < 0.001), and unfavorable DFS/RFS (HR = 2.35, 95% CI 1.90–2.89, P < 0.001) in GC patients. However, CSC markers expression was found to be slightly linked to tumor differentiation (OR = 1.25, 95% CI 1.01–1.55, P = 0.035). Sub-analysis demonstrated a significant positive relationship between most of the individual markers, specially Gli-1, Oct-4, CD44, CD44V6, and CD133, and clinical outcomes as well as the reduced survival, whereas overexpression of Lgr-5, Nanog, and sonic hedgehog (Shh) was not found to be related to the majority of clinical outcomes in GC patients.
Conclusion
The expression of CSC markers is mostly associated with worse outcomes in patients with GC, both overall and individual. The detection of a combined panel of CSC markers might be appropriate as a prognostic stratification marker to predict tumor aggressiveness and poor prognosis in patients with GC, which probably results in identifying novel potential targets for therapeutic approaches.
Keywords: Cancer stem cells, Gastric cancer, Prognostic value, Clinicopathological characteristics, Meta-analysis
Introduction
Gastric cancer (GC) is known as the fifth most common malignancy and the third leading cause of cancer-associated mortality worldwide [1, 2]. It has been reported that nearly 800,000 gastric cancer-related deaths occur annually, with an average 5-year survival rate of less than 30%, which geographically are more frequent in Asian, Eastern European, and South American countries [3]. Although the GC mortality rate has reduced over the last decade due to surgery, chemotherapy, and targeted therapy, the disease burden still remains high with a remarkable unsatisfactory prognosis. Moreover, a high rate of advanced-stage diagnosis, lack of appropriate predictive markers, the progression of recurrence and metastasis, and treatment failure are the key factors that contribute to the poor prognosis of patients with this disease [4].
Of note, biomarkers have become valuable promising tools for improving and optimizing diagnosis, treatment, and prognosis of GC [5]. Nevertheless, the restricted confirmation and controversial prognostic values of the current clinical biomarkers led to qualifying inadequate as robust biomarkers to be implemented in clinical practice for GC patients [6]. Therefore, the molecular pathogenesis of development and progression in GC is still unclear, and more prognosis biomarkers of GC are waiting to be uncovered.
Recently, researchers have focused on identifying and targeting cancer stem cells (CSCs). The heterogeneous phenotype of tumor is considered as a key driver of treatment resistance and cancer recurrence, for which CSCs are considered to be among the major causes of tumor heterogeneity and therapy’s failure [7]. CSCs are defined as a unique subpopulation of cancer cells that possess self-renewal and differentiation potentials, consequently deliberating cancer initiation, invasion, metastasis, relapse, and chemoresistance [8, 9].
Growing evidence supports that overexpression of multiple specific stemness genes in gastrointestinal stem cells may promote the self-renewal capacity of CSCs in GC and are linked to patients’ prognosis [10]. Several cell-surface markers, including CD133, CD44, CD166, and CD24, have been identified as gastric CSC markers [11]. In addition, some pluripotent transcription factors (TFs), including Oct-4, KLF4, MYC, Nanog, and Sox-2 and some intracellular signaling cascades, including Wnt, Sonic hedgehog (Shh), NF-κB, and Notch as well as extracellular factors, are known as essential regulators of CSCs [12]. Recently, many scholars have recognized the relationship of overexpression of CSC-related key markers and possible signal pathways with prognostic and clinical values in gastric carcinoma [13, 14]. However, as a consequence of diversities in study methodology, research participants, and sample size, there are some conflicting opinions on the gastric tumor that need to be addressed. The analysis of these markers may potentially result in the identification of some potential prognostic biomarkers and novel therapeutic targets in GC.
Therefore, we aimed to collect all available data and perform the current meta-analysis on the potential role of CSC-related biomarkers as clinical and prognostic biomarkers in GC patients in order to clarify controversial issues and explain which one of these biomarkers has more clinical importance regarding the quantitative evidence.
Materials and methods
Data sources and literature search strategy
Literature searches, based on the relationship between gastric CSC markers expression and clinical outcomes, were thoroughly performed from PubMed, Scopus, Embase, and ISI Web of Science databases until February 26, 2020, and updated on October 16, 2020. All the searches were restricted to English publications. The search strategy was based on the following main keywords: (neoplasm OR cancer OR tumor OR malignancy OR malignant OR carcinoma) AND (stomach OR gastric) AND (neoplastic stem cell OR neoplastic colony-forming unit OR tumor stem cell OR tumor-initiating cell OR cancer stem cell OR CSC) AND (biomarker OR marker OR prognosis OR prognostic OR diagnosis OR diagnostic OR screening OR detection). The strategy of the literature searching has been presented in Additional file 1: Table S1. The protocol for the current meta-analysis was performed in accordance with the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [15].
Eligibility criteria
Studies were included based on the following criteria: (1) case–control or cohort studies published in English as original studies with available full texts; (2) studies with human gastric tumor; (3) the expression of gastric CSC-relevant markers detected by immunohistochemistry (IHC) in primary tumor tissues instead of serum or other kinds of specimens; (4) studies evaluating the association between the CSC markers expression and overall survival (OS), disease-free survival (DFS)/relapse-free survival (RFS), and disease-specific survival (DSS)/ cancer-specific survival (CSS), and/or clinicopathological features of GC; and (5) hazard ratios (HRs) with 95% confidence intervals (CIs) presented in the text, or availability of data in order to calculate HRs and 95% CIs.
The studies were excluded on the basis of the following criteria: (1) book chapters, reviews, letters, and conference abstracts; (2) studies were not related to the topic of the interest (e.g., when the studies investigated other solid tumors or other diseases); (3) in vitro and animal studies; (4) studies in which participants administered any kind of anticancer therapy such as radiotherapy and chemotherapy, prior to biopsy; and (5) studies with lack of sufficient and useful data.
Study selection and data extraction
All search records were transferred to Endnote software to remove the duplicate files. The eligible studies were identified after the independent screening of the titles and abstracts based on the inclusion criteria by two investigators (MR and RG). Discrepancies were resolved through discussion or by a third investigator. Two independent researchers (MR and RG) extracted prognostic or clinicopathological data from eligible articles in a predefined table. For each of the included articles, the following descriptive data were collected: the name of the first author, country and year of conduction of the study, detection method, age, sex, sample size, CSC marker, case number of different groups, median or mean follow-up times, clinicopathological parameters, cut-off value, and the related survival data. HR and 95% CI of OS, DFS/RFS, and CSS/DSS were taken into account for counting pooled HR. Where HR was not reported, the calculation method was applied to extract HR and 95% CI. The primary outcome was the relationship between the CSC markers expression and OS, DFS/RFS, or CSS/DSS in GC patients. Other outcomes of interest were the relationships between the CSC markers expression and the important clinicopathological parameters of GC. All the extracted data were cross-checked by SV, MS, and LS.
Quality assessment
The quality of all eligible studies was evaluated through the Newcastle–Ottawa Scale (NOS) [16]. Accordingly, the NOS evaluates the quality of studies based on three parameters, i.e., selection, comparability, and exposure or outcome, with a score between 0 to 9. Articles with NOS points above 6 were determined as high-quality studies. Any disagreement was discussed and then resolved by consensus.
Statistical analysis
The associations between gastric CSC markers expression and clinicopathological characteristics, including TNM stage (III/IV vs. I/II), tumor differentiation (poor vs. well/moderate), and lymph node metastasis (positive vs. negative), were evaluated by combining the odds ratios (ORs) and 95% CIs. In the current analysis, an OR > 1 demonstrated a higher possibility of cancer development in GC patients with the CSC markers overexpression. To assess the value of CSC markers overexpression on the prognosis of GC patients, pooled HRs with 95% CI values of survival outcomes, including OS, DFS/RFS, and CSS/DSS, were calculated. HRs were derived from both multivariate and univariate statistical tests by favoring information from multivariate statistics if applicable. When the Kaplan–Meyer curve was presented without declaring HR, it was calculated by Kaplan-Meyer curve according to the method described by Parmar et al. [17]. In this regard, the software GetData Graph Digitizer (http://getdata-graph-digitizer.com/) was utilized to extract survival data from Kaplan-Meyer curves. A pooled HR larger than 1 reflected a poor prognosis in GC patients. The heterogeneity among the included studies was determined through the I2 statistics. Random and fixed-effects models were employed for pooling the data based on the heterogeneity of the included studies. In the presence of considerable heterogeneity (P < 0.05 and/or an I2 statistic > 50%), random-effect models were applied; Otherwise, the fixed-effects models were utilized. Afterward, subgroup analyses on the basis of the expression of individual CSC markers were also employed to examine the possible cause of heterogeneity. Thereafter, the possible publication bias was graphically evaluated through funnel plots and statistically through Egger’s test. All these statistical analyses were conducted using the software Comprehensive Meta-Analysis Version 2.2.064. A two-tailed P < 0.05 defined statistical significance.
Results
Baseline study characteristics
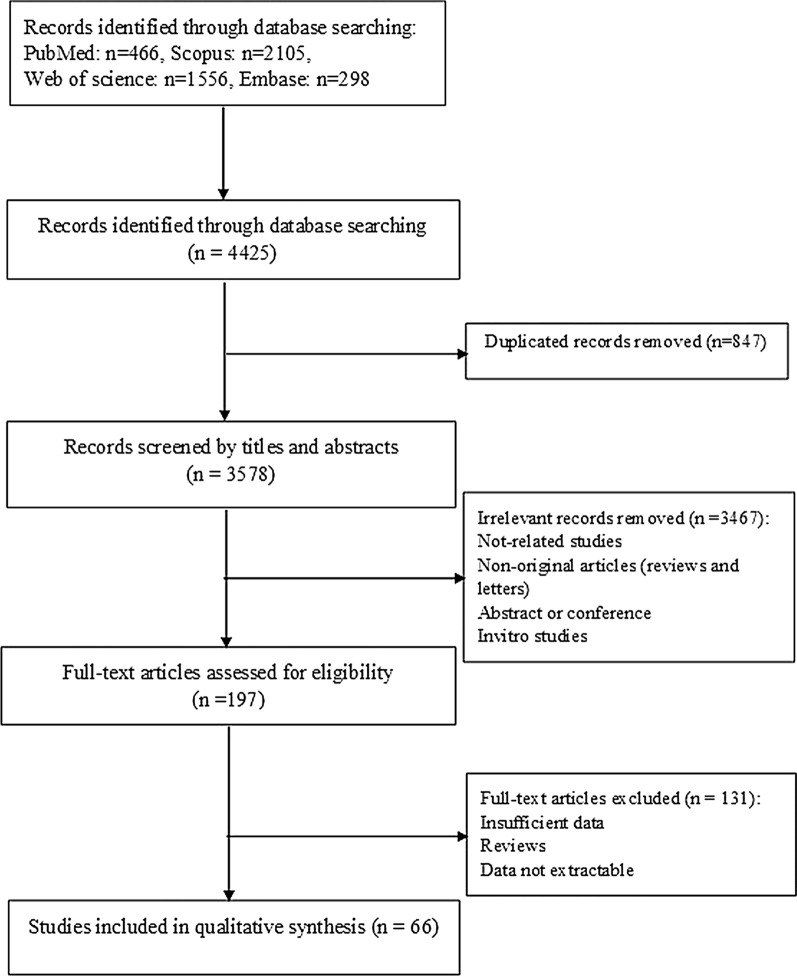
The details of the literature search and selection procedure are presented in a flowchart (Fig. 1). After carefully screening the titles, abstracts, and full text, a total of 66 publications, including 89 studies performed on 11,274 patients, were included for the present meta-analysis according to the inclusion criteria. Table 1 presents the main features of the included papers and patients’ demographics. Notably, all the eligible articles were written in English published between 2002 and 2020, with sample sizes ranging from 40 to 487 participants. According to the NOS quality assessment listed in Table 1, 66 publications were categorized as high quality ranged from 6 to 9. Geographically, most of the articles (n = 38) were carried out in China, while the remaining articles (n = 28) were conducted in other countries (Japan, Korea, Portugal, Netherlands, Germany, Turkey, Thailand, Egypt, Iran, Singapore, and Taiwan). Moreover, in the majority of studies, a large number of participants were male. All 66 articles applied the IHC detection method for analyzing tissue. Notably, 15 publications analyzed the same patient cohorts but by the use of different markers. To account for this purpose, each marker was incorporated in the related pooled analysis, while, for the total number of patients, these studies were only counted once. Among 66 relevant papers with 89 studies, several CSC markers (n = 13) were investigated; of them, 13 studies were performed on CD44s, 13 studies on CD133, 8 studies on Gli-1, 7 studies on Shh, 7 studies on Oct-4, 6 studies on Sox-2, 6 studies on Lgr-5, 6 studies on ALDH1, 5 studies on Bmi-1, 5 studies on CD44V6, 5 studies on CD44V9, 5 studies on CD24, and 3 studies on Nanog. The cut-off values were determined as a score on the basis of the intensity, percentage, or/and the number of positively stained cancer cells (Table 1). Furthermore, 78 studies evaluated the prognostic value of the CSC markers on OS, whilst 18 and 8 investigations assessed the prognostic importance of the markers on DFS/RFS and DSS/CSS, respectively.
Fig. 1.
Flow chart of the literature search and selection procedure based on the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines
Table 1.
Main characteristics of the studies included in the meta-analysis
| Authors year | CSC markers | Country duration | Mean/median age (month) | Sample size (n) | Gender (M/F) | TNM stage | Cut-off value | Follow-up (month) | Outcome | Hazard ratio (HR) | NOS score# |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Wang 2011 [18] |
CD44, CD133 |
Singapore 2000–2004 |
64 | 116 | 76/36 | I–IV |
CD44: 5 score (0–300) CD133: 50 score (0–300) |
110 | OS | E | 8 |
|
Wakamatsu 2012 [19] |
CD44, CD133, ALDH1 |
Japan NR |
65 | 190 | NR | I–IV | > 10% | 83 | OS |
CD44, CD133: R-Multi ALDH1: R-Uni |
6 |
|
Chen 2013 [20] |
CD44, CD133 |
China 2000–2005 |
55 (23 to 84) | 152 | 101/51 | I–IV |
CD44: > 65% CD133: > 50% |
48.6 (1.0–76.7) | OS, DFS |
OS: R-Multi DFS: E |
9 |
|
Cao 2014 [21] |
CD44 |
China 2005–2007 |
60.5 (33–88) | 203 | 140/63 | NR | > 10% | 66 (7–108) | OS | E | 6 |
|
Cao 2014 [22] |
CD44, CD24 |
China 2000–2010 |
64 (32 to 87) | 290 | 221/69 | I–IV | ≥ 30 (0–300) | 41(3–135) | OS | R-Multi | 9 |
|
Jian-Hui 2016 [23] |
CD44, Gli-1, Shh |
China 2006 |
60 | 101 | 62/39 | I–III | ≥ 4 score (0–12) | 90 | OS, DFS |
CD44, Shh: R-Uni Gli: R-Multi |
8 |
|
Zhang 2016 [24] |
CD44, CD133, Sox-2, Oct-4, Gli-1 |
China 2005–2011 |
NR | 101 | NR | I–IV | > 10% | 80 | OS | E | 6 |
|
Kodama 2017 [25] |
CD44, CD44v6, CD44v9 |
Japan 2007–2009 |
70 (38–91) | 123 | 83/40 | I–IV | > 5% | 68 (1–97) | DSS | R-Uni | 7 |
|
Senel 2017 [26] |
CD44, ALDH1 |
Turkey 2011–2015 |
NR | 80 | 48/32 | I–III | > 1 score (0–3) | 60 | DFS | E | 7 |
|
Sun 2017 [27] |
CD44 |
China 2004–2012 |
60 | 200 | 135/65 | I–IV | > 3 score (0–6) | 110 | OS | E | 8 |
|
Tongtawee 2017 [28] |
CD44 |
Thailand 2011–2015 |
61.38 ± 12.39 | 162 | 117/45 | I–IV | > 0 score (0–3) | 60 | OS | R-Multi | 8 |
|
Ryu 2018 [29] |
CD44 |
Korea 1998–2009 |
60.7(27–84) | 143 | 91/52 | NR | > 2 score (0–3) | 45 (0–155) | OS, DFS | R-Multi | 7 |
|
Ibrahim 2019 [30] |
CD44 |
Egypt 2012–2016 |
52.5(41–60) | 40 | 25/15 | I–III | > 10% | 8–36 | OS, DFS | R-Uni | 9 |
|
Xie 2015 [31] |
CD44v6 |
China 2006–2008 |
60 | 208 | 154/54 | I–IV | > 3 score | 80 | OS | R-Multi | 8 |
|
Xu 2017 [32] |
CD44v6 |
China 2006–2013 |
60 | 103 | 69/34 | I–IV | NR | 120 | OS | E | 7 |
|
Yamaguchi 2002 [33] |
CD44v6 |
Japan 1984–1993 |
NR | 201 | NR | I–IV | NR | 144 | OS | E | 6 |
|
Zheng 2017 [34] |
CD44v6 |
China 2010–2015 |
65 (43–80) | 49 | 39/10 | NR | NR | 33 (1–73) | DSS | R | 9 |
|
Hirata 2013 [35] |
CD44v9 |
Japan 2008–2010 |
67.5 (46–90) | 65 | 56/9 | NR | 3.58 ± 7.74% | 32 (1–36) | RFS | R-Multi | 7 |
|
Yamakawa 2017 [36] |
CD44v9 |
Japan 2011–2012 |
70 | 103 | 79/24 | I–IV | ≥ 4 score | 61.0 | RFS | R-Multi | 8 |
|
Go 2016 [37] |
CD44v9 |
Korea 1999–2007 |
24–85 | 333 | 218/115 | I–III | > 0 score (0–3) | 120 | OS | E | 8 |
|
Songun 2005 [38] |
CD44v9 |
Netherlands 1989–1993 |
64.7 (31–84) | 300 | 181 /119 | I–IV | ≥ 5% | 120 | OS | E | 6 |
|
Ishigami 2010 [39] |
CD133 |
Japan 2001–2003 |
65 (40 to 85) | 97 | 69/28 | I–IV | containing at least one CD133 positive cell | 60 | OS | E | 8 |
|
Yu 2010 [40] |
CD133 |
China 2004–2009 |
62.0 (29–83) | 99 | 69/30 | I–IV | NR | 26.76 ± 17.02 | OS | R-Multi | 8 |
|
Zhao 2010 [41] |
CD133 |
China NR |
58.1 (18–85) | 336 | 274/62 | I–IV | ≥ 5 score (0–12) | 120 | OS | E | 7 |
|
Lee 2012 [42] |
CD133 |
Korea 2001–2005 |
61.5 (29–89) | 100 | 71/29 | II–III | ≥ 6 score (0–12) | 46.9 (0–115) | OS, DFS | R-Multi | 8 |
|
Hashimoto 2014 [43] |
CD133 |
Japan 2004–2006 |
66 ± 11 | 189 | 133/56 | I–IV | > 5% | 60 | OS | E | 7 |
|
Zhou 2015 [44] |
CD133, LGR-5 |
China 2004–2006 |
59.6 (23–88) | 261 | 172/89 | I–IV | > 2 score (0–12) | 43.9 (0–110) | OS | R-Multi | 7 |
|
Lu 2017 [45] |
CD133 |
China 2010–2011 |
58.1 ( 28–78) | 236 | 159/77 | I–III | ≥ 3 score (0–12) | 48.6 (9–72) | OS | R-Multi | 7 |
|
Attia 2019 [46] |
CD133 |
Egypt 2012–2015 |
54.44 (24–81) | 77 | 45/32 | I–IV | > 3.5 score (0–6) | NR | RFS | R | 6 |
|
Liu 2019 [47] |
CD133, ALDH1 |
China 2012–2017 |
53 (32–76) | 91 | 69/22 | III | ≥ 5 score (0–12) | 27 (1–60) | OS, DFS |
OS: E DFS:R-multi |
7 |
|
Matsuoka 2012 [48] |
Sox-2, Oct-4, Nanog |
Japan NR |
NR | 290 | NR | I–IV | ≥ 5 score (0–5) | 120 | OS | R-Uni | 6 |
|
Li 2015 [49] |
SOX-2, Oct-4, Nanog |
China 2008–2009 |
55 (28–78) | 69 | 41/18 | I–III | ≥ 5 score (0–5) | 35(6–60) | OS, DFS | E | 8 |
|
Yang 2017 [50] |
Sox-2, ALDH1 |
China 2010–2013 |
63 (29–82) | 122 | 100/22 | I–IV | > 5% | 26 (1 to 75) | OS |
Sox-2: R-Multi ALDH1: R-Uni |
8 |
|
Zhang 2010 [51] |
Sox-2 |
China 2004 |
57.8 ( 29–79) | 50 | 35/15 | I–IV | > 10% | 1–50 | OS | E | 7 |
|
Camilo 2014 [52] |
Sox-2 |
Portugal 1988–2010 |
66.5 (24–89) | 201 | 124/77 | I–IV | > 5% | 250 | OS | E | 8 |
|
Kong 2014 [53] |
Oct-4 |
China NR |
60 | 158 | 104/54 | I–IV | > 0 score (0–3) | 60 | OS | R-Multi | 6 |
|
Jiang 2016 [54] |
Oct-4 |
China 2001–2010 |
62 (30–85) | 412 | 284/128 | I–IV | ≥ 4 score | 60 | OS | R-Multi | 7 |
| Javanbakht 2017 [55] | Oct-4 |
Iran 2010–2014 |
59.3 (37–85) | 40 | 16/24 | I–IV | > 25% | 50 | OS | R-Multi | 6 |
|
El-Guindy 2019 [56] |
Oct-4 |
Egypt 2015–2016 |
18–70 | 45 | 28/17 | I–III | ≥ 6 score (0–9) | 24 | OS, DFS | E | 6 |
|
Li 2014 [57] |
ALDH1 |
China 2005–2008 |
57.0 ( 22–82) | 216 | 140/76 | I–III | ≥ 2 score (0–3) | 27 (4–82) | OS, RFS | R-Multi | 8 |
|
41Lu 2018 [58] |
ALDH1 |
China 2011–2012 |
60 | 232 | 148/84 | I–III | > 2 score (0–12) | 48.7 (10–83) | OS | R-Multi | 7 |
|
Simon 2012 [59] |
LGR-5 |
Germany 1997–2009 |
68 + _11.4 | 487 | 304/183 | I–IV | NR | 60.9 (14.3–129.9) | OS | E | 6 |
|
Bu 2013 [60] |
LGR-5 |
China 2002–2007 |
61 (22–87) | 257 | 185/72 | I–IV | ≥ 3 score (0–6) | 60 | OS | R-Uni | 8 |
|
Xi 2014 [61] |
LGR-5 |
China 1999–2004 |
59.6 (24–86) | 318 | 254/59 | I–IV | ≥ 2 score (0–6) | 120 | OS | R-Multi | 7 |
|
Choi 2017 [62] |
LGR-5 |
Korea 2004–2006 |
60 | 456 | 312/144 | I–IV | NR | NR | CSS | R-Multi | 6 |
|
Liu 2019 [63] |
LGR-5 |
China 2009–2014 |
60 (33 to 85) | 100 | 68/32 | I–III | > 6 score (0–12) | 60 | OS | E | 8 |
|
Liu 2008 [64] |
Bmi-1 |
China 1999–2002 |
60 | 146 | 92/54 | I–IV | ≥ 10% | 48 | OS | R-Multi | 7 |
|
Zhang 2010 [65] |
Bmi-1 |
China NR |
60 | 75 | 50/25 | I–IV | ≥ 10% | 110 | OS | E | 7 |
|
Yang 2011 [66] |
Bmi-1 |
China 2004–2005 |
60 | 219 | 162/57 | I–IV | ≥ 20% | 29.57 (1.90–78.10) | DSS | E | 7 |
|
Wu 2016 [67] |
Bmi-1 |
China NR |
60 | 352 | 236/116 | I–IV | NR | 120 | OS | E | 6 |
|
Lu 2012 [68] |
Bmi-1 |
China 1993–2006 |
55 | 309 | 98/211 | NR | > 5% | 67.8 (2–200) | OS | E | 7 |
|
Lin 2012 [69] |
Nanog |
China NR |
50 | 105 | 71/34 | I–IV | > 2 score (0–12) | 60 | OS | R-Multi | 7 |
|
Chou 2007 [70] |
CD24 |
Taiwan 1995–1997 |
73 (40–99) | 103 | 75/28 | I–IV | > 10% | 86.5 (1–120) | OS | R-Multi | 6 |
|
Fujikuni 2014 [71] |
CD24 |
Japan 2001–2008 |
65 | 119 | 73/46 | I–III | > 1% | 60 | OS | E | 6 |
|
Darwish 2004 [72] |
CD24 |
Korea NR |
54.3 | 300 | 200/100 | I–IV | NR | 53 (1–72) | OS | R-Multi | 8 |
|
Bektas 2010 [73] |
CD24 |
Turkey 2001–2009 |
61.5 (30–84) | 93 | 59/34 | I–IV | > 10% | 36 (0–100) | OS | E | 8 |
|
Hu 2017 [74] |
Gli-1, Shh |
China NR |
60 | 90 | 53/37 | I–IV | ≥ 6 score (0–12) | 80 | OS | E | 6 |
|
Shao 2017 [75] |
Gli-1 |
China 2008–2010 |
60 | 67 | 50/17 | I–IV | ≥ 5 score (0–12) | 60 | OS | R-Multi | 9 |
|
Tang 2018 [76] |
Gli-1 |
China 2009–2010 |
60 | 90 | 68/22 | I–IV | > 3 score (0–9) | 80 | OS | R-Multi | 7 |
|
Ke 2020 [77] |
Gli-1, Shh |
China 2010–2013 |
60 | 128 | 128/50 | I–III | > 3 score | 120 | OS | R-Multi | 8 |
|
Wang 2014 [78] |
Gli-1 |
China 2005–2007 |
63 (46–83) | 121 | 92/29 | I–IV | ≥ 10% | 30 ( 5–60) | OS | R-Multi | 7 |
|
Yao 2019 [79] |
Gli-1 |
China 2010–2012 |
60 | 57 | 40/17 | I–IV | ≥ 5 score (0–12) | 60 | OS | E | 9 |
|
Ertao 2016 [80] |
Shh |
China 2004–2005 |
60 | 117 | 71/46 | I–IV | > 10% | 47.6 (3–114) | OS | R-Multi | 8 |
|
Kim 2012 [81] |
Shh |
Korea 2004–2007 |
NR | 319 | NR | I–IV | > 3 score (0–4) | 80 | OS | E | 6 |
|
Yoo 2011 [82] |
Shh |
Korea 2002–2004 |
60 | 178 | 124/49 | I–IV | NR | 60 | OS | E | 7 |
|
Niu 2014 [83] |
Shh |
China 2008–2009 |
54.37 (22–75) | 113 | 73/40 | I–IV | ≥ 2 score (0–3) | 43.6 (6–78) | OS | R-Multi | 7 |
HR: Hazard ratio; OS: Overall survival; RFS: Relapse-free survival; DSS: Disease-specific survival; DFS: Disease-free survival; CSS: Cancer-specific survival; NR: Not reported; IHC: Immunohistochemistry; E: Estimated by survival curve; R: Reported; R-Uni: Univariate analysis reported by article; R-Multi: Multivariate analysis reported by article; # Study quality was evaluated according to the Newcastle–Ottawa Scale (range, 1–9)
Association between cancer stem cell (CSC) markers expression and clinicopathological features
Table 2 exhibits findings of the relationship between CSC markers expression and the clinicopathological parameters. Overall analyses of the markers indicated that overexpression of CSC markers is significantly associated with TNM stage (OR = 2.19, 95% CI 1.84–2.61, P = 0.013) and lymph node metastasis (OR = 1.76, 95% CI 1.54–2.02, P < 0.001) with considerable heterogeneity (I2 = 80.74%, Ph < 0.001 and I2 = 73.57%, Ph < 0.001), respectively. No significant association was found between CSC markers and tumor differentiation (OR = 1.25, 95% CI 1.01–1.55, P = 0.035; I2 = 82.38%, Ph < 0.001). Thereafter, we assessed the association between the expression of individual CSC markers and clinicopathological features. As shown in Table 2, the expressions of ALDH1 (OR = 3.66, 95% CI 1.75–7.64, P < 0.001; I2 = 74.19%, Ph = 0.009), Bmi-1 (OR = 2.85, 95% CI 1.04–7.81, P = 0.041; I2 = 88.46%, Ph < 0.001), CD133 (OR = 2.67, 95% CI 1.84–3.89, P < 0.001; I2 = 54.01%, Ph = 0.016), CD44 (OR = 2.74, 95% CI 1.39–5.38, P = 0.003; I2 = 83.96%, Ph < 0.001), CD44V6 (OR = 2.50, 95% CI 1.22–5.14, P = 0.012; I2 = 70.46%, Ph = 0.034), CD44V9 (OR = 5.39, 95% CI 2.70–10.77, P < 0.001; I2 = 7.26%, Ph = 0.29), Gli-1 (OR = 4.00, 95% CI 1.58–10.13, P = 0.003; I2 = 79.50%, Ph < 0.001), and Oct-4 (OR = 2.25, 95% CI 1.09–4.66, P = 0.028; I2 = 81.91%, Ph < 0.001) were associated with TNM stage. Moreover, it was found that only CD44s expression is statistically linked to tumor differentiation (OR = 2.82, 95% CI 1.50–5.30, P = 0.001; I2 = 75.38%, Ph < 0.001). Additionally, there was a relationship between the expressions of Oct-4 (OR = 2.70, 95% CI 1.09–6.68, P = 0.031; I2 = 88.63%, Ph < 0.001), Bmi-1 (OR = 2.26, 95% CI 1.19–4.26, P = 0.012; I2 = 76.82%, Ph = 0.002), CD133 (OR = 1.85, 95% CI 1.22–2.79, P = 0.003; I2 = 65.05%, Ph = 0.001), CD44 (OR = 1.64, 95% CI 1.13–2.38, P = 0.009; I2 = 56.83%, Ph = 0.01), CD44V6 (OR = 2.26, 95% CI 1.46–3.51, P < 0.001; I2 = 25.64%, Ph = 0.25), CD24 (OR = 1.40, 95% CI 1.04–1.89, P = 0.026; I2 = 20.17%, Ph = 0.28), Gli-1 (OR = 3.04, 95% CI 1.62–5.71, P = 0.001; I2 = 51.59%, Ph = 0.08), and Sox-2 (OR = 1.96, 95% CI 1.40–2.73, P < 0.001; I2 = 0.00%, Ph = 0.42) and lymph node metastasis. However, no clear association was identified between some CSC markers, including LGR-5, Nanog, or Shh, and the clinicopathological features (all; P > 0.05).
Table 2.
Main results of pooled HRs in the meta-analysis
| Study number | Sample size | OR/HR (95% CI) | P value | z | Heterogeneity | Publication bias (Eggers test) | |||
|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | Ph | T-value | Pbias | ||||||
| Overall TNM stage (III/IV vs I/II) | 70 | 11,901 | 2.19 (1.84–2.61) | P < 0.001 | 8.83 | 80.74 | P < 0.001 | 2.40 | 0.018 |
| Overall tumor differentiation (poor vs well/moderate) | 59 | 9251 | 1.25 (1.01–1.55) | 0.035 | 2.10 | 82.38 | P < 0.001 | 1.84 | 0.07 |
| Overall lymph node metastasis (Yes vs No) | 76 | 13,172 | 1.76 (1.54–2.02) | 0.00 | 8.23 | 73.57 | P < 0.001 | 2.37 | 0.019 |
| Overall OS | 78 | 13,482 | 1.65 (1.54–1.77) | 0.00 | 14.00 | 56.74 | P < 0.001 | 3.70 | P < 0.001 |
| Overall DFS/RFS | 18 | 1788 | 2.35 (1.90–2.89) | P < 0.001 | 7.96 | 46.93 | 0.015 | 1.63 | 0.12 |
| Overall CSS/ DSS | 8 | 1462 | 1.69 (1.33–2.15) | P < 0.001 | 4.32 | 49.04 | 0.056 | 1.00 | 0.35 |
| CD44s | |||||||||
| TNM stage (III/IV vs I/II) | 9 | 1331 | 2.74 (1.39–5.38) | 0.003 | 2.93 | 83.96 | P < 0.001 | 1.55 | 0.163 |
| Tumor differentiation (poor vs well/moderate) | 9 | 1177 | 2.82 (1.50–5.30) | 0.001 | 3.23 | 75.38 | P < 0.001 | 0.71 | 0.49 |
| Lymph node metastasis (Yes vs No) | 11 | 1638 | 1.64 (1.13–2.38) | 0.009 | 2.62 | 56.83 | 0.010 | 1.65 | 0.13 |
| OS | 11 | 1669 | 1.97 (1.55–2.50) | P < 0.001 | 5.61 | 54.99 | 0.014 | 1.62 | 0.13 |
| DFS/RFS | 5 | 516 | 2.74 (1.80–4.17) | P < 0.001 | 4.69 | 42.23 | 0.14 | – | – |
| CSS/DSS | 2 | 246 | 2.59 (1.32–5.06) | 0.005 | 2.78 | 0.00 | 0.65 | – | – |
| CD44V6 | |||||||||
| TNM stage (III/IV vs I/II) | 3 | 311 | 2.50 (1.22–5.14) | 0.012 | 2.51 | 70.46 | 0.034 | 0.40 | 0.75 |
| Tumor differentiation (poor vs well/moderate) | 5 | 684 | 0.93 (0.35–2.42) | 0.88 | − 0.14 | 86.80 | P < 0.001 | 0.55 | 0.61 |
| Lymph node metastasis (Yes vs No) | 5 | 684 | 2.26 (1.46–3.51) | P < 0.001 | 3.65 | 25.64 | 0.25 | 1.68 | 0.19 |
| OS | 3 | 512 | 1.81 (1.29–2.53) | 0.001 | 3.45 | 0.00 | 0.82 | 2.34 | 0.25 |
| CSS/DSS | 3 | 295 | 3.29 (1.46–7.43) | 0.004 | 2.87 | 61.63 | 0.10 | – | – |
| CD44V9 | |||||||||
| TNM stage (III/IV vs I/II) | 2 | 637 | 5.39 (2.70–10.77) | P < 0.001 | 4.78 | 7.26 | 0.29 | – | – |
| Tumor differentiation (poor vs well/moderate) | 3 | 559 | 0.98 (0.37–2.58) | 0.98 | − 0.025 | 84.72 | 0.001 | 14.65 | 0.043 |
| Lymph node metastasis (Yes vs No) | 3 | 559 | 1.52 (0.91–2.52) | 0.10 | 1.61 | 40.74 | 0.18 | 8.64 | 0.073 |
| OS | 2 | 619 | 1.22 (0.96–1.54) | 0.08 | 1.69 | P < 0.001 | 0.74 | – | – |
| DFS/RFS | 2 | 168 | 8.60 (1.70–43.57) | 0.009 | 2.61 | 77.51 | 0.035 | – | – |
| CSS/DSS | 2 | 246 | 1.67 (0.52–5.35) | 0.38 | 0.86 | 0.00 | 0.48 | – | – |
| CD133 | |||||||||
| TNM stage (III/IV vs I/II) | 11 | 1853 | 2.67 (1.84–3.89) | P < 0.001 | 5.16 | 54.01 | 0.016 | 0.25 | 0.80 |
| Tumor differentiation (poor vs well/moderate) | 10 | 1677 | 0.87 (0.56–1.34) | 0.53 | − 0.62 | 70.18 | P < 0.001 | 2.09 | 0.06 |
| Lymph node metastasis (Yes vs No) | 11 | 1847 | 1.85 (1.22–2.79) | 0.003 | 2.93 | 65.05 | 0.001 | 0.99 | 0.34 |
| OS | 12 | 1939 | 1.74 (1.52–2.01) | P < 0.001 | 7.87 | 0.00 | 0.55 | 1.83 | 0.09 |
| DFS/RFS | 3 | 343 | 2.59 (1.74–3.85) | P < 0.001 | 4.69 | 0.00 | 0.64 | – | – |
| Sox-2 | |||||||||
| TNM stage (III/IV vs I/II) | 5 | 732 | 1.37 (0.65–2.90) | 0.40 | 0.83 | 77.22 | 0.002 | 1.55 | 0.21 |
| Tumor differentiation (poor vs well/moderate) | 2 | 119 | 1.21 (0.57–2.59) | 0.60 | 0.51 | 23.37 | 0.25 | – | – |
| Lymph node metastasis (Yes vs No) | 5 | 732 | 1.96 (1.40–2.73) | P < 0.001 | 3.99 | 0.00 | 0.42 | 0.84 | 0.46 |
| OS | 6 | 804 | 1.73 (1.37–2.18) | P < 0.001 | 4.63 | 0.00 | 0.78 | 0.59 | 0.58 |
| DFS/RFS | 1 | 69 | 1.31 (0.68–2.49) | 0.41 | 0.82 | 0.00 | 1 | – | – |
| Oct-4 | |||||||||
| TNM stage (III/IV vs I/II) | 6 | 1014 | 2.25 (1.09–4.66) | 0.028 | 2.19 | 81.91 | P < 0.001 | 0.79 | 0.47 |
| Tumor differentiation (poor vs well/moderate) | 5 | 724 | 1.97 (0.86–4.50) | 0.10 | 1.60 | 75.47 | 0.003 | 0.53 | 0.62 |
| Lymph node metastasis (Yes vs No) | 6 | 1014 | 2.70 (1.09–6.68) | 0.031 | 2.15 | 88.63 | P < 0.001 | 1.15 | 0.31 |
| OS | 7 | 1086 | 1.87 (1.48–2.35) | 0.00 | 5.37 | 4.88 | 0.38 | 0.21 | 0.83 |
| DFS/RFS | 2 | 114 | 2.18 (1.06–4.48) | 0.023 | 2.13 | 11.13 | 0.28 | – | – |
| ALDH1 | |||||||||
| TNM stage (III/IV vs I/II) | 4 | 760 | 3.66 (1.75–7.64) | 0.001 | 3.45 | 74.19 | 0.009 | 1.31 | 0.31 |
| Tumor differentiation (poor vs well/moderate) | 3 | 539 | 2.73 (0.43–17.19) | 0.28 | 1.07 | 93.25 | P < 0.001 | 4.6 | 0.13 |
| Lymph node metastasis (Yes vs No) | 4 | 760 | 2.50 (0.96–6.45) | 0.058 | 1.89 | 88.08 | P < 0.001 | 0.09 | 0.93 |
| OS | 5 | 851 | 1.65 (1.32–2.05) | P < 0.001 | 4.53 | 0.00 | 0.49 | 2.60 | 0.08 |
| DFS/RFS | 2 | 307 | 1.49 (0.73–3.00) | 0.27 | 1.10 | 56.39 | 0.13 | – | – |
| LGR-5 | |||||||||
| TNM stage (III/IV vs I/II) | 5 | 1392 | 1.31 (0.46–3.74) | 0.60 | 0.51 | 94.11 | P < 0.001 | 0.58 | 0.6 |
| Tumor differentiation (poor vs well/moderate) | 5 | 1423 | 1.04 (0.50–2.13) | 0.91 | 0.10 | 87.79 | P < 0.001 | 0.36 | 0.73 |
| Lymph node metastasis (Yes vs No) | 6 | 1879 | 1.21 (0.70–2.09) | 0.48 | 0.70 | 84.70 | P < 0.001 | 0.01 | 0.98 |
| OS | 5 | 1879 | 1.26 (0.97–1.63) | 0.075 | 1.78 | 60.45 | 0.039 | 1.03 | 0.37 |
| CSS/DSS | 1 | 456 | 1.02 (0.67–1.54) | 0.90 | 0.11 | 0.00 | 1.00 | – | – |
| Bmi-1 | |||||||||
| TNM stage (III/IV vs I/II) | 4 | 792 | 2.85 (1.04–7.81) | 0.041 | 2.04 | 88.46 | P < 0.001 | 1.07 | 0.36 |
| Tumor differentiation (poor vs well/moderate) | 4 | 792 | 1.14 (0.70–1.85) | 0.57 | 0.55 | 51.72 | 0.10 | 0.09 | 0.93 |
| Lymph node metastasis (Yes vs No) | 5 | 1101 | 2.26 (1.19–4.26) | 0.012 | 2.51 | 76.82 | 0.002 | 1.13 | 0.33 |
| OS | 4 | 882 | 1.32 (0.77–2.27) | 0.30 | 1.06 | 84.49 | P < 0.001 | 1.57 | 0.25 |
| CSS/DSS | 1 | 219 | 1.97 (1.35–2.87) | P < 0.001 | 3.52 | 0.00 | 1.00 | – | – |
| Nanog | |||||||||
| TNM stage (III/IV vs I/II) | 3 | 464 | 1.34 (0.78–2.31) | 0.27 | 1.08 | 0.86 | P < 0.001 | 0.49 | 0.70 |
| Tumor differentiation (poor vs well/moderate) | 2 | 174 | 1.89 (0.06–55.82) | 0.71 | 0.37 | 93.05 | P < 0.001 | – | – |
| Lymph node metastasis (Yes vs No) | 3 | 464 | 1.49 (0.73–3.02) | 0.26 | 1.10 | 44.43 | 0.16 | 0.71 | 0.60 |
| OS | 3 | 464 | 1.59 (0.68–3.77) | 0.28 | 1.07 | 80.97 | 0.005 | 0.93 | 0.52 |
| DFS/RFS | 1 | 69 | 2.09 (0.92–4.70) | 0.075 | 1.77 | 0.00 | 1 | – | – |
| CD24 | |||||||||
| TNM stage (III/IV vs I/II) | 5 | 905 | 1.31 (0.87–1.98) | 0.18 | 1.32 | 43.46 | 0.13 | 0.91 | 0.42 |
| Tumor differentiation (poor vs well/moderate) | 2 | 393 | 1.10 (0.18–6.46) | 0.91 | 0.10 | 90.02 | 0.002 | – | – |
| Lymph node metastasis (Yes vs No) | 5 | 905 | 1.40 (1.04–1.89) | 0.026 | 2.23 | 20.17 | 0.28 | 0.46 | 0.67 |
| OS | 5 | 905 | 1.73 (1.25–2.4) | 0.001 | 4.10 | 35.46 | 0.18 | 2.71 | 0.07 |
| Gli-1 | |||||||||
| TNM stage (III/IV vs I/II) | 6 | 614 | 4.00 (1.58–10.13) | 0.003 | 2.93 | 79.50 | P < 0.001 | 1.07 | 0.34 |
| Tumor differentiation (poor vs well/moderate) | 4 | 403 | 1.66 (0.81–3.40) | 0.16 | 1.39 | 57.08 | 0.07 | 0.93 | 0.44 |
| Lymph node metastasis (Yes vs No) | 5 | 493 | 3.04 (1.62–5.71) | 0.001 | 3.47 | 51.59 | 0.08 | 0.60 | 0.59 |
| OS | 8 | 776 | 1.75 (1.34–2.31) | 0.00 | 4.06 | 7.79 | 0.37 | 0.14 | 0.88 |
| DFS/RFS | 1 | 101 | 3.40 (1.30–8.7) | 0.012 | 2.52 | 0.00 | 1 | – | – |
| Shh | |||||||||
| TNM stage (III/IV vs I/II) | 7 | 1096 | 2.19 (0.97–4.94) | 0.056 | 1.90 | 86.32 | P < 0.001 | 3.12 | 0.026 |
| Tumor differentiation (poor vs well/moderate) | 5 | 687 | 1.02 (0.24–4.30) | 0.97 | 0.037 | 90.31 | P < 0.001 | 1.63 | 0.20 |
| Lymph node metastasis (Yes vs No) | 7 | 1096 | 1.14 (0.54–2.39) | 0.73 | 0.34 | 85.02 | P < 0.001 | 0.48 | 0.64 |
| OS | 7 | 1096 | 1.27 (0.78–2.05) | 0.32 | 0.99 | 76.23 | P < 0.001 | 0.72 | 0.49 |
| DFS/RFS | 1 | 101 | 2.75 (1.15–6.54) | 0.02 | 2.28 | 0.00 | 1 | – | – |
Ph: The p-value of heterogeneity; P bias: The p-value of Egger test for assessing publication bias; OS: Overall survival; DFS: Disease-free survival; RFS: Relapse-free survival; CSS: Cancer-specific survival; DSS: Disease-specific survival
Association between cancer stem cell (CSC) markers expression and overall survival (OS)
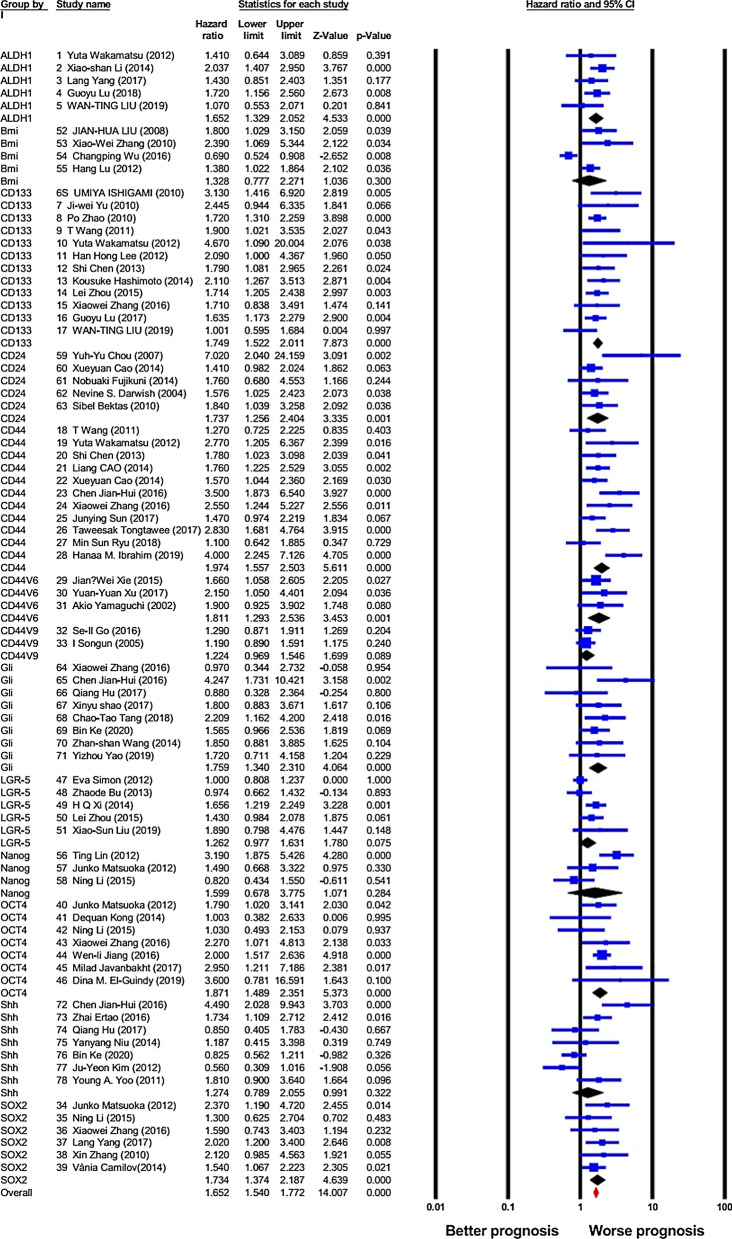
A total of 78 studies provided adequate information to evaluate the link between CSC markers and OS in patients with GC, and as presented in Fig. 2 and Table 2, overexpression of overall CSC markers notably predicted worse OS in GC patients (HR = 1.65, 95% CI 1.54–1.77, P < 0.001), with a rather moderate heterogeneity (I2 = 56.74%, P < 0.001). Consequently, a random-effect model was utilized. Afterward, the relationship between expression of individual CSC markers and OS was studied. Accordingly, the obtained results showed that overexpression of ALDH1 (HR = 1.65, 95% CI 1.32–2.05, P < 0.001; I2 = 0.00%, Ph = 0.49), CD133 (HR = 1.74, 95% CI 1.52–2.01, P < 0.001; I2 = 0.00%, Ph = 0.55), CD24 (HR = 1.73, 95% CI 1.25–2.4, P < 0.001; I2 = 33.46%, Ph = 0.18), CD44 (HR = 1.97, 95% CI 1.55–2.50, P < 0.001; I2 = 54.99%, Ph = 0.014), CD44V6 (HR = 1.81, 95% CI 1.29–2.53, P = 0.001; I2 = 0.00%, Ph = 0.82), Gli-1 (HR = 1.75, 95% CI 1.34–2.31, P < 0.001; I2 = 7.79%, Ph = 0.37), Oct-4 (HR = 1.87, 95% CI 1.48–2.35, P < 0.001; I2 = 4.88%, Ph = 0.38), and Sox-2 (HR = 1.73, 95% CI 1.37–2.18, P < 0.001; I2 = 0.00%, Ph = 0.78) are positively linked with worse OS in GC patients. However, data analysis indicated no statistically meaningful association between the overexpression of Bmi-1 (HR = 1.32, 95% CI 0.77–2.27, P = 0.3; I2 = 84.49%, Ph < 0.001), CD44V9 (HR = 1.22, 95% CI 0.96–1.54, P = 0.08; I2 = 0.00%, Ph = 0.74), LGR-5 (HR = 1.26, 95% CI 0.97–1.63, P = 0.07; I2 = 60.45%, Ph = 0.039), Nanog (HR = 1.59, 95% CI 0.67–3.77, P = 0.28; I2 = 80.97%, Ph = 0.005) or Shh (HR = 1.27, 95% CI 0.78–2.05, P = 0.32; I2 = 76.23%, Ph < 0.001) and OS.
Fig. 2.
Forest plot showing the results of the association between cancer stem cell (CSC) markers expression and overall survival (OS) in gastric cancer (GC)
Association between cancer stem cell (CSC) markers expression and disease-free survival (DFS)/relapse-free survival (RFS)
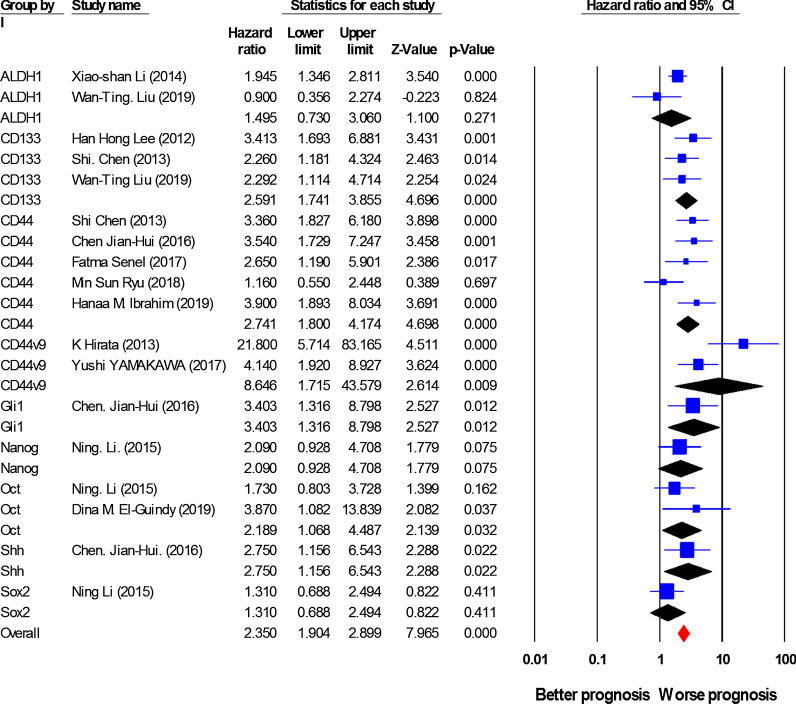
18 studies including 1788 patients investigated the relationship between expression of CSC markers and DFS/RFS. As shown in Fig. 3 and Table 2, the overall pooled HR indicated a considerable elevated risk of disease progression or recurrence in the cases with high expression of CSC markers (HR = 2.35, 95% CI 1.90–2.89, P < 0.001), with a rather slight heterogeneity (I2 = 46.93%, P = 0.015). Analyses for individual CSC markers indicated that high expression of CSC markers, including CD133 (HR = 2.59, 95% CI 1.74–3.85, P < 0.001; I2 = 0.00%, Ph = 0.64), CD44 (HR = 2.74, 95% CI 1.80–4.17, P < 0.001; I2 = 42.23%, Ph = 0.14), CD44V9 (HR = 8.60, 95% CI 1.70–43.57, P = 0.009; I2 = 77.51%, Ph = 0.035), and Oct-4 (HR = 2.18, 95% CI 1.06–4.48, P = 0.023; I2 = 4.88%, Ph = 0.38) is associated with a poor DFS/RFS. Additionally, a limited number of publications reported the association of Gli-1 (n = 1, HR = 3.40, 95% CI 1.30–8.70, P = 0.012) and Shh (n = 1, HR = 2.75, 95% CI 1.15–6.54, P = 0.02) with DFS/RFS. However, data analysis showed no remarkable effects of overexpression of ALDH1 (HR = 1.49, 95% CI 0.73–3, P = 0.27; I2 = 0.00%, Ph = 0.49), Nanog (HR = 2.09, 95% CI 0.92–4.7, P = 0.075; I2 = 80.97%, Ph = 0.005) or Sox-2 (HR = 1.31, 95% CI 0.68–2.49, P = 0.41; I2 = 0.00%, Ph = 0.78) on DFS/RFS. Furthermore, in this study, the relationships between overexpression of Bmi-1, CD24, CD44v6 or LGR-5 and DFS/RFS were not investigated due to the lack of sufficient information.
Fig. 3.
Forest plot showing the results of the association between cancer stem cell (CSC) markers expression and disease-free survival/ relapse-free survival (DFS/RFS) in gastric cancer (GC)
Association between cancer stem cell (CSC) markers expression and cancer-specific survival (CSS)/ Disease-specific survival (DSS)
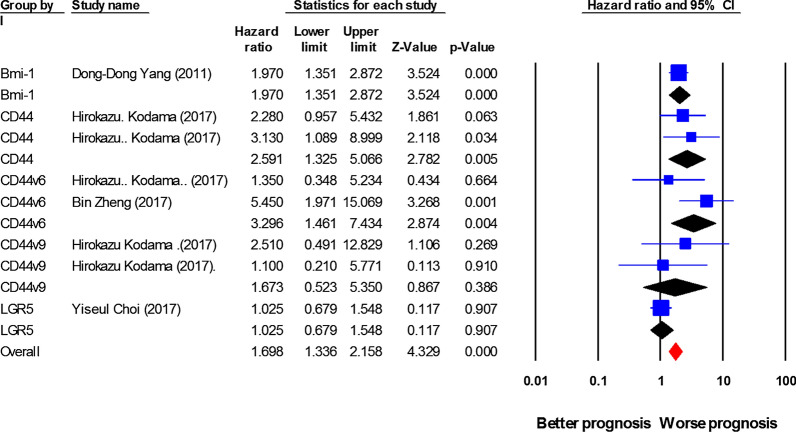
HRs for CSC markers were presented in 8 studies, involving 1462 cases. The pooled HR indicated a considerable prognostic importance of CSC markers overexpression in GC patients CSS/DSS prediction (HR = 1.69, 95% CI 1.33–2.15, P < 0.001, Fig. 4 and Table 2), with a slight heterogeneity (I2 = 49.04%, P = 0.056). From one study, more than one HR was extracted for CSS/DSS, because the expression of multiple CSC markers was investigated [25]. A subgroup analysis on the basis of the expression of CSC markers indicated that although Bmi-1 (HR = 1.97, 95% CI 1.35–2.87, P < 0.001), CD44s (HR = 2.59, 95% CI 1.32–5.06, P = 0.005; I2 = 61.63%, Ph = 0.1), and CD44V6 (HR = 3.29, 95% CI 1.46–7.43, P = 0.004; I2 = 61.63%, Ph = 0.1) are significantly associated with GC patients CSS/DSS, CD44V9 (HR = 1.67, 95% CI 0.52–5.35, P = 0.38; I2 = 0.00%, Ph = 0.48) and LGR-5 (HR = 1.02, 95% CI 0.67–1.54, P = 0.9) are not associated. Moreover, the associations between ALDH1, CD133, CD24, Oct-4, Gli-1, Shh, Sox-2, Nanog, or LGR-5 expression and CSS/DSS were not studied owing to the inadequacy of data.
Fig. 4.
Forest plot showing the results of the association between cancer stem cell (CSC) markers expression and cancer-specific survival/ Disease-specific survival (CSS/ DSS) in gastric cancer (GC)
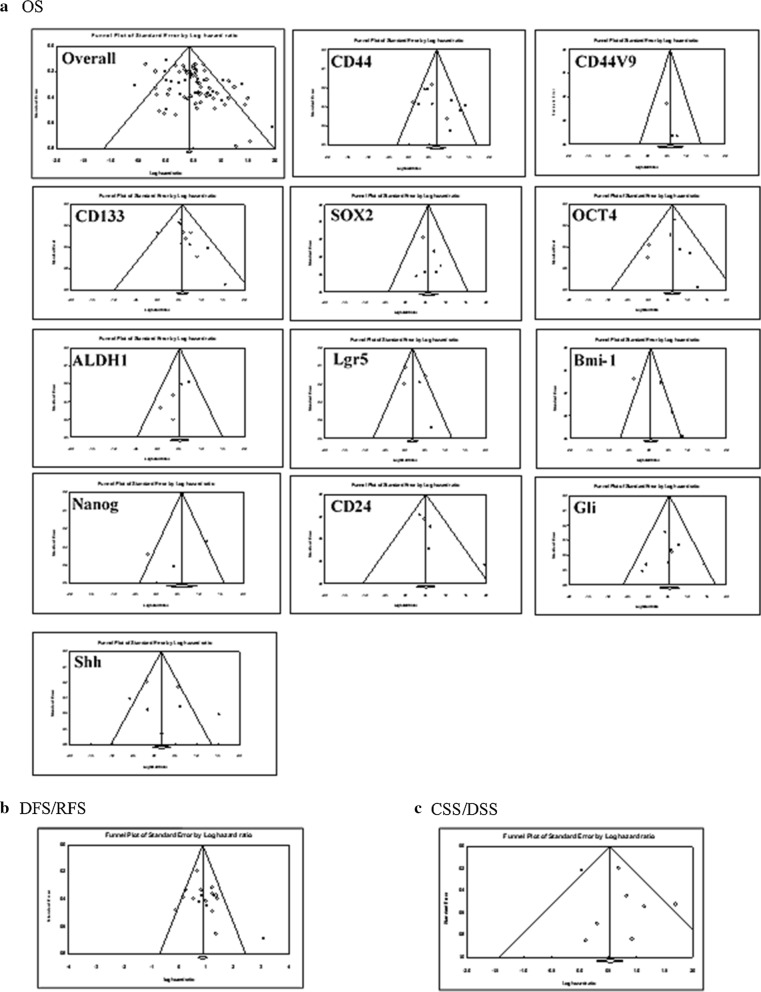
Publication Bias
In the current meta-analysis, the presence of the publication bias in the eligible studies was evaluated for OS, DFS/RFS, and DSS/CSS using funnel plot analysis (Fig. 5) and Egger’s test (Table 2). Graphically, the funnel plots showed that asymmetry might be present in the graph of the studies concerning overall OS, but probably not in DFS/RFS and DSS/CSS studies (Fig. 5), proposing the possible existence of a publication bias about OS. Subsequently, Egger’s tests were conducted to investigate the bias more precisely. The studies regarding the expression of overall CSC markers and OS (Table 2) demonstrated a significant publication bias as evaluated through Egger’s test (t-value = 3.7, P < 0.001). Then, the publication bias was evaluated for individual CSC markers and OS, in which Egger’s test and funnel plot graphs revealed nonsignificant publication bias for each CSC marker. However, we did not perform the publication bias analysis for the relationship between the expression of individual CSCs markers and DFS/RFS or DSS/CSS because of the limited number of eligible studies.
Fig. 5.
Funnel plots for publication bias test with 95%confidence limits. a Cancer stem cell (CSC) markers associated with overall survival (OS); b CSC markers associated with disease-free survival/ relapse-free survival (DFS/RFS); c CSC markers associated with cancer-specific survival/ Disease-specific survival (CSS/DSS)
Discussion
This review provided the most comprehensive meta-analysis of gastric CSC biomarkers and recognized clinicopathological and prognostic significances for CSC markers. Our meta-analysis presented robust evidence for the association between CSC biomarkers expression and GC prognosis by enrolling 11,274 GC participants, emphasizing the potential clinical applicability of CSC biomarkers in GC. Principally, we attempted to address the study heterogeneity and publication bias.
Generally, we identified a strong association between higher levels of CSC biomarkers and TNM stage, lymph node metastasis, poor OS, DFS/RFS, and CSS/DSS, suggesting their important roles in prognosis and patient stratification. These findings suggest that gastric CSC markers may assist clinicians and decision-makers in evaluating GC status after surgery. However, the overall expression of CSC markers had a slight association with tumor differentiation of GC cells. Based on the obtained findings, CSC markers are likely to have a more key role in the relapse of GC (HR = 2.35) compared to death resulted from GC (HR = 1.69). However, because of the limited sample sizes regarding CSS/DSS and DFS/RFS, our findings should be interpreted with caution. Data involving clinical value and prognostic significance of overall CSC markers expression were characterized by partially high heterogeneity, and, to specify the positive staining for each marker, a considerable variability within the cut-off thresholds utilized in the various studies was identified. Variability of patients’ characteristics and the different antibodies used for the detection of CSC markers might lead to heterogeneity among these investigations. Additionally, publication bias was detected in the pooled HR for the OS and overall CSC markers expression. However, analyses based on the individual expression of CSC markers were free of any publication bias.
Gastric CSC markers have been suggested to interact with each other and with several signaling pathways, which were found to be associated with aggressive phenotype [84, 85]. Accumulating evidence has displayed that CSCs can promote growth, migration, angiogenesis, and metastasis of gastric tumor cells, which may support the association between the expression of CSC markers and clinical outcomes [86, 87]. CSCs are a minor subpopulation within the total cancer cells, making their identification in the heterogeneous masses of cells challenging. The appropriate approach to detect and target CSCs has been determined to be the utilization of cell-surface markers in different cancer types [88]. However, a lack of uniform expression of the already used markers might limit their advantages for CSCs detection, and, notably, inconsistencies still remain regarding the ideal markers panel to detect gastric CSCs. CD44, a transmembrane glycoprotein, is the first identified cell-surface marker used commonly for the isolation of CSCs [89], and subsequently, the associations of CD24, CD44v6, CD44v9, CD133, LGR-5, and cytosolic ALDH1 with the clinicopathological parameters of cancers have been investigated in various studies [19, 90–92]. In GC patient-derived xenografts, tumor biopsies, and cell lines, the fraction of cells with overexpression of these CSC markers displays self-renewal, tumorigenic, multilineage differentiation, and chemoresistance abilities, proposing that these may be robust CSC biomarkers [12, 93]. Consistently, our results also indicate that a positive expression of cell-surface CSC markers, including CD24, CD133, CD44s, CD44V6, and CD44V9, as well as cytosolic ALDH1 marker, can strongly predict the important clinicopathological parameters along with worse prognostic outcomes in GC cases. Importantly, considering that the related parameters do not overlap, the detection of a combined panel of CSC markers is likely to provide a more precise predictive potential for GC patients. However, CD44s was the only CSC marker linked to tumor differentiation, suggesting involving of CD44s in malignant progression of GC. In addition, data regarding the expression of Lgr-5 showed no association with clinicopathological and prognostic values of patients with the disease. Moreover, a few references exist concerning the link between the expression of CD44V9 and GC clinical outcomes that our results indicate no association between its expression and poor OS and CSS/DSS.
The other specific stemness-related marker types in gastric cancer are transcription factors (TFs) implicated in self-renewal and pluripotency. Key stem cell TFs such as Sox-2, Oct-4, and Nanog have been demonstrated to be overexpressed in CSCs [94]. Our results suggest that, while a high expression of Oct-4 is associated with clinicopathological features that can reduce the OS and DFS/RFS of GC patients, overexpression of Nanog has no association with prognostic and clinicopathological values of patients. The results of a research by Li et al. [49] were also in agreement with our findings, suggesting that Oct-4 might be a more useful prognostic factor for relapse or distant metastasis after operation compared with Nanog and Sox-2. In GC, Sox-2 function is still accompanied by some controversies; some studies reported the link of its overexpression with a more aggressive feature, worse prognosis, and chemoresistance [48, 95], while some other studies have demonstrated the opposite [49]. We found a positive relationship between Sox-2 overexpression and lymph node metastasis as well as OS, but not with DFS/RFS. Bmi-1 is another TF that has been found to importantly regulate the self-renewal capacity of both normal and tumor stem cells [96]. It has been demonstrated that Bmi-1 overexpression might considerably associate with a worse OS in breast cancer cases [97]. However, in our sub-analysis, Bmi-1 did not affect the OS of GC cases. Instantly, lymph node metastasis, TNM stage, and worse CSS/DSS in GC patients were associated with the overexpression of Bmi-1, representing the role of Bmi-1 in the death resulted from GC.
Additionally, many intracellular signaling pathways have been cleared to be involved in the regulation of CSCs [93]. The sonic hedgehog (Shh) signaling cascade is one of these pathways that is occurred through the binding of Shh ligands to transmembrane receptor Patched 1 (PTCH1), which allows the dissociation and conformational change of Smoothened (Smo) receptor, eventually resulting in the activation of three zinc finger Gli transcription factors (Gli-1, Gli-2, and Gli-3) [98]. Gli-1 appears to play a pivotal role in the maintenance of tumor cells with stemness characteristics. In GC, Gli-1 expression has been reported to be positively linked to a more aggressive tumor phenotype [79]. Similarly, our analysis indicates that Gli-1 overexpression not only promotes higher TNM stage and lymph node metastasis strongly but also reduces OS and DFS/RFS in GC patients. On the other hand, the expression of Shh has also been found to contribute to epithelial-to-mesenchymal transition (EMT) in pancreatic adenocarcinoma cell lines [99] as well as lymphatic metastasis in bladder cancer [100]. However, our analyses display that overexpression of Shh is not associated with clinicopathological values and OS in GC patients. Nevertheless, based on only one study [23], its high expression was linked to a poor DFS/RFS in GC patients.
However, our large meta-analysis sheds light on the clinicopathological and prognostic roles of gastric CSC markers; there are several potential limitations that must be considered as well as some results that should be interpreted with caution. Firstly, this meta-analysis was not performed based on the randomized controlled studies with a prospective design, which would have made this study more susceptible to information and selection biases. Secondly, the cut-off values defining CSC markers overexpression in the eligible studies were not based on a universal standard, possibly affecting the findings of this meta-analysis. Third, since most of the included articles in the current study were performed in Asian countries, a potential population selection bias may be produced. Fourth, the present meta-analysis was restricted to the articles published in English, which might be along with selection bias. Fifth, for papers without providing HR with 95% CI directly, we estimated the HRs through Kaplan–Meier curves, which possibly reduces the credibility of the findings. Sixth, partially high heterogeneity was identified in most analyses. Differences in research methodology and race might affect the heterogeneity. Finally, the publication bias was detected for OS and the overexpression of overall CSC markers, therefore likely reducing the reliability of the association between CSCs and worse prognosis. The majority of the studies prefer to selectively publish positive findings, potentially resulting in publication bias. Considering all these limitations, further multicenter prospective investigations on the basis of the standardized methodology are needed to validate the potential of the gastric CSC markers in the prediction of patients’ outcomes.
Conclusion
In conclusion, findings of our comprehensive meta-analysis reveal a notable role of CSC markers, including cell surface markers, TFs, and components of Shh signaling pathway, in predicting poor clinical outcomes of patients with GC. Previous meta-analyses evaluated only some of these associations and cleared that some CSC markers have prognostic significance for OS of GC patients [101, 102]. However, to the best of our knowledge, this is the first meta-analysis that exclusively included 13 CSC markers with a large sample size, which made the findings more robust and powerful, and on the other side, systematically assessed the clinicopathological and prognostic values of CSC markers, overall and individually, among gastric cancer patients. Although all of the detected CSC markers are not predictors of worse outcomes, most can potentially be known as the prognostic biomarkers. By analyzing the eligible studies, it was cleared that Gli-1, Oct-4, CD44s, CD44V6, and CD133 have strong prognostic values. Our meta-analysis suggests applying a combined panel of CSC markers overexpression for the prediction of gastric cancer patients OS, DFS/RFS, and CSS/DSS and the stratification of different gastric cancer patients. However, due to some certain limitations, various analyses showed relatively inconsistent results, of which careful selection of CSC markers and the standardized methodology are possibly considered as the fundamental ones to optimize the accuracy of CSC markers as prognostic and predictive clinical factors in gastric cancer.
Supplementary Information
Additional file 1: Table S1. Search strategy and syntax in different databases based on the expression of cancer stem cell markers in gastric cancer.
Acknowledgements
Not applicable.
Abbreviations
- GC
Gastric cancer
- CSC
Cancer stem cell
- HRs
Hazard ratios
- ORs
Odds ratios
- CIs
Confidence intervals
- IHC
Immunohistochemistry
- OS
Overall survival
- DFS
Disease-free survival
- CSS
Cancer-specific survival
- Shh
Sonic hedgehog
- TFs
Transcription factors
- NOS
Newcastle–Ottawa Scale
- PTCH1
Patched 1 receptor
- Smo
Smoothened receptor
- EMT
Epithelial-to-mesenchymal transition
- RFS
Relapse-free survival
- DSS
Disease-specific survival
Authors’ contributions
MR and ZM designed the review. MR and RG performed the literature search, literature selection, and data extraction. SV, MS, and LS checked the data extraction. MR and SV statistically analyzed the obtained data. MR and ZM wrote the manuscript. All authors contributed to the preparation of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Iran University of Medical Sciences (Grant # 99–3-28–19475).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All of the current study data were searched systematically and were used by reference citation, and all of the authors consent to publication.
Competing interests
The authors declare that there are no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mahdieh Razmi, Email: Razmi_mahdyeh@yahoo.com, Email: razmi.mahdieh@ut.ac.ir.
Zahra Madjd, Email: Zahra.madjd@yahoo.com, Email: majdjabari.z@iums.ac.ir.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-021-01840-z.
References
- 1.Chandra R, Balachandar N, Wang S, Reznik S, Zeh H, Porembka M. The changing face of gastric cancer: epidemiologic trends and advances in novel therapies. Cancer Gene Ther. 2020 doi: 10.1038/s41417-020-00234-z. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Pellino A, Riello E, Nappo F, Brignola S, Murgioni S, Djaballah SA, et al. Targeted therapies in metastatic gastric cancer: Current knowledge and future perspectives. World J Gastroenterol. 2019;25(38):5773–5788. doi: 10.3748/wjg.v25.i38.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrillo A, Pompella L, Tirino G, Pappalardo A, Laterza MM, Caterino M, et al. Perioperative treatment in resectable gastric cancer: current perspectives and future directions. Cancers (Basel) 2019;11(3):399. doi: 10.3390/cancers11030399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner AD, Lordick F, Grabsch HI, Terashima M, Terada M, Yoshikawa T, et al. Multidisciplinary management of stage II-III gastric and gastro-oesophageal junction cancer. Eur J Cancer. 2020;124:67–76. doi: 10.1016/j.ejca.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Gao Y, Xi H, Wei B, Cui J, Zhang K, Li H, et al. Association between liquid biopsy and prognosis of gastric cancer patients: a systematic review and meta-analysis. Front Oncol. 2019;9:1222. doi: 10.3389/fonc.2019.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16(1):41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata M, Hoque MO. Targeting cancer stem cells: a strategy for effective eradication of cancer. Cancers (Basel) 2019;11(5):732. doi: 10.3390/cancers11050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba H, Ishimoto T. Targeting cancer stem cells in gastric cancer. Gastrointest Cancer Targets Therapy. 2014;4:123–129. doi: 10.2147/GICTT.S48999. [DOI] [Google Scholar]
- 10.Huang T, Song X, Xu D, Tiek D, Goenka A, Wu B, Sastry N, Hu B, Cheng SY. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10(19):8721–8743. doi: 10.7150/thno.41648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bekaii-Saab T, El-Rayes B. Identifying and targeting cancer stem cells in the treatment of gastric cancer. Cancer. 2017;123(8):1303–1312. doi: 10.1002/cncr.30538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5(1):8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yiming L, Yunshan G, Bo M, Yu Z, Tao W, Gengfang L, et al. CD133 overexpression correlates with clinicopathological features of gastric cancer patients and its impact on survival: a systematic review and meta-analysis. Oncotarget. 2015;6(39):42019–42027. doi: 10.18632/oncotarget.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandra V, Lee YM, Gupta U, Mittal B, Kim JJ, Rai R. Quantitative assessment of CD44 genetic variants and cancer susceptibility in Asians: a meta-analysis. Oncotarget. 2016;7(45):74286–74302. doi: 10.18632/oncotarget.10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 17.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Ong CW, Shi J, Srivastava S, Yan B, Cheng CL, et al. Sequential expression of putative stem cell markers in gastric carcinogenesis. Br J Cancer. 2011;105(5):658–665. doi: 10.1038/bjc.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakamatsu Y, Sakamoto N, Oo HZ, Naito Y, Uraoka N, Anami K, Sentani K, Oue N, Yasui W. Expression of cancer stem cell markers ALDH1, CD44 and CD133 in primary tumor and lymph node metastasis of gastric cancer. Pathol Int. 2012;62(2):112–119. doi: 10.1111/j.1440-1827.2011.02760.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Hou JH, Feng XY, Zhang XS, Zhou ZW, Yun JP, Chen YB, Cai MY. Clinicopathologic significance of putative stem cell marker, CD44 and CD133, in human gastric carcinoma. J Surg Oncol. 2013;107(8):799–806. doi: 10.1002/jso.23337. [DOI] [PubMed] [Google Scholar]
- 21.Cao L, Hu X, Zhang J, Liang P, Zhang Y. CD44(+) CD324(-) expression and prognosis in gastric cancer patients. J Surg Oncol. 2014;110(6):727–733. doi: 10.1002/jso.23690. [DOI] [PubMed] [Google Scholar]
- 22.Cao X, Cao D, Jin M, Jia Z, Kong F, Ma H, Wang Y, Jiang J. CD44 but not CD24 expression is related to poor prognosis in non-cardia adenocarcinoma of the stomach. BMC Gastroenterol. 2014;14:157. doi: 10.1186/1471-230X-14-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jian-Hui C, Er-Tao Z, Si-Le C, Hui W, Kai-Ming W, Xin-Hua Z, Chuang-Qi C, Shi-Rong C, Yu-Long H. CD44, Sonic Hedgehog, and Gli1 Expression are prognostic biomarkers in gastric cancer patients after radical resection. Gastroenterol Res Pract. 2016;2016:1013045. doi: 10.1155/2016/1013045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Hua R, Wang X, Huang M, Gan L, Wu Z, et al. Identification of stem-like cells and clinical significance of candidate stem cell markers in gastric cancer. Oncotarget. 2016;7(9):9815–9831. doi: 10.18632/oncotarget.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama H, Murata S, Ishida M, Yamamoto H, Yamaguchi T, Kaida S, et al. Prognostic impact of CD44-positive cancer stem-like cells at the invasive front of gastric cancer. Br J Cancer. 2017;116(2):186–194. doi: 10.1038/bjc.2016.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senel F, Kokenek Unal TD, Karaman H, Inanc M, Aytekin A. Prognostic value of cancer stem cell markers CD44 and ALDH1/2 in gastric cancer cases. Asian Pac J Cancer Prev. 2017;18(9):2527–2531. doi: 10.22034/APJCP.2017.18.9.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun JY, Sun BC, Zhu DW, Zhao XL, Zhang YH, Dong XY, et al. HMGA2 regulates CD44 expression to promote gastric cancer cell motility and sphere formation. Am J Cancer Res. 2017;7(2):260–274. [PMC free article] [PubMed] [Google Scholar]
- 28.Tongtawee T, Wattanawongdon W, Simawaranon T, Kaewpitoon S, Kaengpenkae S, Jintabanditwong N, et al. Expression of cancer stem cell marker CD44 and its polymorphisms in patients with chronic gastritis, precancerous gastric lesion, and gastric cancer: A cross-sectional multicenter study in Thailand. Biomed Res Int. 2017;2017:4384823. doi: 10.1155/2017/4384823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryu MS, Park HJ, Moon CM, Kim S-E, Jung H-K, Shim K-N, Jung S-A, Cho MS. Expression of CD44 according to clinicopathologic characteristics of gastric cancer. Ewha Med J. 2018;41(3):63–74. doi: 10.12771/emj.2018.41.3.63. [DOI] [Google Scholar]
- 30.Ibrahim HM, AbdElbary AM, Mohamed SY, Elwan A, Abdelhamid MI, Ibrahim A. Prognostic value of cyclin D1 and CD44 expression in gastric adenocarcinoma. J Gastrointest Cancer. 2019;50(3):370–379. doi: 10.1007/s12029-018-0079-2. [DOI] [PubMed] [Google Scholar]
- 31.Xie JW, Chen PC, Zheng CH, Li P, Wang JB, Lin JX, et al. Evaluation of the prognostic value and functional roles of CD44v6 in gastric cancer. J Cancer Res Clin Oncol. 2015;141(10):1809–1817. doi: 10.1007/s00432-015-1964-8. [DOI] [PubMed] [Google Scholar]
- 32.Xu YY, Guo M, Yang LQ, Zhou F, Yu C, Wang A, et al. Regulation of CD44v6 expression in gastric carcinoma by the IL-6/STAT3 signaling pathway and its clinical significance. Oncotarget. 2017;8(28):45848–45861. doi: 10.18632/oncotarget.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi A, Goi T, Yu J, Hirono Y, Ishida M, Iida A, et al. Expression of CD44v6 in advanced gastric cancer and its relationship to hematogenous metastasis and long-term prognosis. J Surg Oncol. 2002;79(4):230–235. doi: 10.1002/jso.10082. [DOI] [PubMed] [Google Scholar]
- 34.Zheng B, Ni CH, Chen H, Wu WD, Guo ZH, Zhu Y, Zheng W, Chen C. New evidence guiding extent of lymphadenectomy for esophagogastric junction tumor: Application of Ber-Ep4 Joint with CD44v6 staining on the detection of lower mediastinal lymph node micrometastasis and survival analysis. Medicine (Baltimore) 2017;96(14):e6533. doi: 10.1097/MD.0000000000006533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirata K, Suzuki H, Imaeda H, Matsuzaki J, Tsugawa H, Nagano O, Asakura K, Saya H, Hibi T. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br J Cancer. 2013;109(2):379–386. doi: 10.1038/bjc.2013.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamakawa Y, Kusuhara M, Terashima M, Kinugasa Y, Sugino T, Abe M, et al. CD44 variant 9 expression as a predictor for gastric cancer recurrence: immunohistochemical and metabolomic analysis of surgically resected tissues. Biomed Res. 2017;38(1):41–52. doi: 10.2220/biomedres.38.41. [DOI] [PubMed] [Google Scholar]
- 37.Go SI, Ko GH, Lee WS, Kim RB, Lee JH, Jeong SH, Lee YJ, Hong SC, Ha WS. CD44 variant 9 serves as a poor prognostic marker in early gastric cancer, but not in advanced gastric cancer. Cancer Res Treat. 2016;48(1):142–152. doi: 10.4143/crt.2014.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Songun I, Litvinov SV, van de Velde CJ, Pals ST, Hermans J, van Krieken JH. Loss of Ep-CAM (CO17-1A) expression predicts survival in patients with gastric cancer. Br J Cancer. 2005;92(9):1767–1772. doi: 10.1038/sj.bjc.6602519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishigami S, Ueno S, Arigami T, Uchikado Y, Setoyama T, Arima H, et al. Prognostic impact of CD133 expression in gastric carcinoma. Anticancer Res. 2010;30(6):2453–2457. [PubMed] [Google Scholar]
- 40.Yu JW, Zhang P, Wu JG, Wu SH, Li XQ, Wang ST, Lu RQ, Ni XC, Jiang BJ. Expressions and clinical significances of CD133 protein and CD133 mRNA in primary lesion of gastric adenocacinoma. J Exp Clin Cancer Res. 2010;29:141. doi: 10.1186/1756-9966-29-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao P, Li Y, Lu Y. Aberrant expression of CD133 protein correlates with Ki-67 expression and is a prognostic marker in gastric adenocarcinoma. BMC Cancer. 2010;10:218. doi: 10.1186/1471-2407-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HH, Seo KJ, An CH, Kim JS, Jeon HM. CD133 expression is correlated with chemoresistance and early recurrence of gastric cancer. J Surg Oncol. 2012;106(8):999–1004. doi: 10.1002/jso.23178. [DOI] [PubMed] [Google Scholar]
- 43.Hashimoto K, Aoyagi K, Isobe T, Kouhuji K, Shirouzu K. Expression of CD133 in the cytoplasm is associated with cancer progression and poor prognosis in gastric cancer. Gastric Cancer. 2014;17(1):97–106. doi: 10.1007/s10120-013-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L, Yu L, Feng ZZ, Gong XM, Cheng ZN, Yao N, Wang DN, Wu SW. Aberrant expression of markers of cancer stem cells in gastric adenocarcinoma and their relationship to vasculogenic mimicry. Asian Pac J Cancer Prev. 2015;16(10):4177–4183. doi: 10.7314/APJCP.2015.16.10.4177. [DOI] [PubMed] [Google Scholar]
- 45.Lu GY, Zhou L, Song WQ, Wu SW, Zhu B, Wang DN. Expression of ORAOV1, CD133 and WWOX correlate with metastasis and prognosis in gastric adenocarcinoma. Int J Clin Exp Pathol. 2017;10(8):8916–8924. [PMC free article] [PubMed] [Google Scholar]
- 46.Attia S, Atwan N, Arafa M, Shahin RA. Expression of CD133 as a cancer stem cell marker in invasive gastric carcinoma. Pathologica. 2019;111(1):18–23. doi: 10.32074/1591-951X-51-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu WT, Liu WB, Gao M, Zhang YY, Gu KS. Expression of ALDH1A1 and CD133 is associated with the prognosis and effect of different chemotherapeutic regimens in gastric cancer. Oncol Lett. 2019;18(5):4573–4582. doi: 10.3892/ol.2019.10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuoka J, Yashiro M, Sakurai K, Kubo N, Tanaka H, Muguruma K, Sawada T, Ohira M, Hirakawa K. Role of the stemness factors sox2, oct3/4, and nanog in gastric carcinoma. J Surg Res. 2012;174(1):130–135. doi: 10.1016/j.jss.2010.11.903. [DOI] [PubMed] [Google Scholar]
- 49.Li N, Deng W, Ma J, Wei B, Guo K, Shen W, Zhang Y, Luo S. Prognostic evaluation of Nanog, Oct4, Sox2, PCNA, Ki67 and E-cadherin expression in gastric cancer. Med Oncol. 2015;32(1):433. doi: 10.1007/s12032-014-0433-6. [DOI] [PubMed] [Google Scholar]
- 50.Yang L, Xu JF, Kang Q, Li AQ, Jin P, Wang X, et al. Predictive value of stemness factor Sox2 in gastric cancer is associated with tumor location and stage. PLoS ONE. 2017;12(1):e0169124. doi: 10.1371/journal.pone.0169124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Yu H, Yang Y, Zhu R, Bai J, Peng Z, et al. SOX2 in gastric carcinoma, but not Hath1, is related to patients' clinicopathological features and prognosis. J Gastrointest Surg. 2010;14(8):1220–1226. doi: 10.1007/s11605-010-1246-3. [DOI] [PubMed] [Google Scholar]
- 52.Camilo V, Celestino R, Castro P, Vieira J, Teixeira MR, Carneiro F, et al. Immunohistochemical molecular phenotypes of gastric cancer based on SOX2 and CDX2 predict patient outcome. BMC Cancer. 2014;14:753. doi: 10.1186/1471-2407-14-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong D, Su G, Zha L, Zhang H, Xiang J, Xu W, Tang Y, Wang Z. Coexpression of HMGA2 and Oct4 predicts an unfavorable prognosis in human gastric cancer. Med Oncol. 2014;31(8):130. doi: 10.1007/s12032-014-0130-5. [DOI] [PubMed] [Google Scholar]
- 54.Jiang WL, Zhang PF, Li GF, Dong JH, Wang XS, Wang YY. Oct-4 is associated with gastric cancer progression and prognosis. Onco Targets Ther. 2016;9:517–522. doi: 10.2147/OTT.S90031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Javanbakht M, Akhavanmoghadam J, Talaei AJ, Aghyani M, Mozafari M, Khedmat L, Mohebbi M. Differential expression of two genes Oct-4 and MUC5AC associates with poor outcome in patients with gastric cancer. Clin Exp Pharmacol Physiol. 2017;44(11):1099–1105. doi: 10.1111/1440-1681.12840. [DOI] [PubMed] [Google Scholar]
- 56.El-Guindy DM, Wasfy RE, Abdel Ghafar MT, Ali DA, Elkady AM. Oct4 expression in gastric carcinoma: association with tumor proliferation, angiogenesis and survival. J Egypt Natl Canc Inst. 2019;31:3. doi: 10.1186/s43046-019-0005-0. [DOI] [PubMed] [Google Scholar]
- 57.Li XS, Xu Q, Fu XY, Luo WS. ALDH1A1 overexpression is associated with the progression and prognosis in gastric cancer. BMC Cancer. 2014;14:705. doi: 10.1186/1471-2407-14-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu G, Wang X, Wang Y, Cheng Z, Zhou L. Value of CagA, HER2, ALDH1, and KiSS-1 in predicting metastasis and prognosis for gastric adenocarcinoma. Int J Clin Exp Pathol. 2018;11(7):3628–3637. [PMC free article] [PubMed] [Google Scholar]
- 59.Simon E, Petke D, Boger C, Behrens HM, Warneke V, Ebert M, Rocken C. The spatial distribution of LGR5+ cells correlates with gastric cancer progression. PLoS ONE. 2012;7(4):e35486. doi: 10.1371/journal.pone.0035486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bu Z, Zheng Z, Zhang L, Li Z, Sun Y, Dong B, et al. LGR5 is a promising biomarker for patients with stage I and II gastric cancer. Chin J Cancer Res. 2013;25(1):79–89. doi: 10.3978/j.issn.1000-9604.2013.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xi HQ, Cai AZ, Wu XS, Cui JX, Shen WS, Bian SB, et al. Leucine-rich repeat-containing G-protein-coupled receptor 5 is associated with invasion, metastasis, and could be a potential therapeutic target in human gastric cancer. Br J Cancer. 2014;110(8):2011–2020. doi: 10.1038/bjc.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi Y, Park J, Ko YS, Kim Y, Pyo JS, Jang BG, et al. FOXO1 reduces tumorsphere formation capacity and has crosstalk with LGR5 signaling in gastric cancer cells. Biochem Biophys Res Commun. 2017;493(3):1349–1355. doi: 10.1016/j.bbrc.2017.09.163. [DOI] [PubMed] [Google Scholar]
- 63.Liu XS, Lin XK, Mei Y, Ahmad S, Yan CX, Jin HL, et al. Regulatory T cells promote overexpression of Lgr5 on gastric cancer cells via TGF-beta1 and confer poor prognosis in gastric cancer. Front Immunol. 2019;10:1741. doi: 10.3389/fimmu.2019.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu JH, Song LB, Zhang X, Guo BH, Feng Y, Li XX, Liao WT, Zeng MS, Huang KH. Bmi-1 expression predicts prognosis for patients with gastric carcinoma. J Surg Oncol. 2008;97(3):267–272. doi: 10.1002/jso.20934. [DOI] [PubMed] [Google Scholar]
- 65.Zhang XW, Sheng YP, Li Q, Qin W, Lu YW, Cheng YF, et al. BMI1 and Mel-18 oppositely regulate carcinogenesis and progression of gastric cancer. Mol Cancer. 2010;9:40. doi: 10.1186/1476-4598-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang DD, Cui BB, Sun LY, Zheng HQ, Huang Q, Tong JX, Zhang QF. The co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinoma. Cell Biochem Biophys. 2011;61(3):703–710. doi: 10.1007/s12013-011-9229-x. [DOI] [PubMed] [Google Scholar]
- 67.Wu C, Zheng X, Li X, Fesler A, Hu W, Chen L, et al. Reduction of gastric cancer proliferation and invasion by miR-15a mediated suppression of Bmi-1 translation. Oncotarget. 2016;7(12):14522–14536. doi: 10.18632/oncotarget.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu H, Sun HZ, Li H, Cong M. The clinicopathological significance of Bmi-1 expression in pathogenesis and progression of gastric carcinomas. Asian Pac J Cancer Prev. 2012;13(7):3437–3441. doi: 10.7314/APJCP.2012.13.7.3437. [DOI] [PubMed] [Google Scholar]
- 69.Lin T, Ding YQ, Li JM. Overexpression of Nanog protein is associated with poor prognosis in gastric adenocarcinoma. Med Oncol. 2012;29(2):878–885. doi: 10.1007/s12032-011-9860-9. [DOI] [PubMed] [Google Scholar]
- 70.Chou YY, Jeng YM, Lee TT, Hu FC, Kao HL, Lin WC, Lai PL, Hu RH, Yuan RH. Cytoplasmic CD24 expression is a novel prognostic factor in diffuse-type gastric adenocarcinoma. Ann Surg Oncol. 2007;14(10):2748–2758. doi: 10.1245/s10434-007-9501-x. [DOI] [PubMed] [Google Scholar]
- 71.Fujikuni N, Yamamoto H, Tanabe K, Naito Y, Sakamoto N, Tanaka Y, et al. Hypoxia-mediated CD24 expression is correlated with gastric cancer aggressiveness by promoting cell migration and invasion. Cancer Sci. 2014;105(11):1411–1420. doi: 10.1111/cas.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darwish NS, Kim MA, Chang MS, Lee HS, Kim YI, Kim WH. Prognostic significance of CD24 expression in gastric carcinoma. Cancer Res Treat. 2004;36(5):298–302. doi: 10.4143/crt.2004.36.5.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bektas S, Bahadir B, Ucan BH, Ozdamar SO. CD24 and galectin-1 expressions in gastric adenocarcinoma and clinicopathologic significance. Pathol Oncol Res. 2010;16(4):569–577. doi: 10.1007/s12253-010-9248-8. [DOI] [PubMed] [Google Scholar]
- 74.Hu Q, Hou YC, Huang J, Fang JY, Xiong H. Itraconazole induces apoptosis and cell cycle arrest via inhibiting Hedgehog signaling in gastric cancer cells. J Exp Clin Cancer Res. 2017;36(1):50. doi: 10.1186/s13046-017-0526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao X, Kuai X, Pang Z, Zhang L, Wu L, Xu L, Zhou C. Correlation of Gli1 and HER2 expression in gastric cancer: Identification of novel target. Sci Rep. 2018;8(1):397. doi: 10.1038/s41598-017-17435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang CT, Liang Q, Yang L, Lin XL, Wu S, Chen Y, Zhang XT, Gao YJ, Ge ZZ. RAB31 targeted by MiR-30c-2-3p regulates the GLI1 signaling pathway, affecting gastric cancer cell proliferation and apoptosis. Front Oncol. 2018;8:554. doi: 10.3389/fonc.2018.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ke B, Wang XN, Liu N, Li B, Wang XJ, Zhang RP, Liang H. Sonic Hedgehog/Gli1 signaling pathway regulates cell migration and invasion via induction of epithelial-to-mesenchymal transition in gastric cancer. J Cancer. 2020;11(13):3932–3943. doi: 10.7150/jca.42900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang ZS, Shen Y, Li X, Zhou CZ, Wen YG, Jin YB, Li JK. Significance and prognostic value of Gli-1 and Snail/E-cadherin expression in progressive gastric cancer. Tumour Biol. 2014;35(2):1357–1363. doi: 10.1007/s13277-013-1185-1. [DOI] [PubMed] [Google Scholar]
- 79.Yao Y, Zhou D, Shi D, Zhang H, Zhan S, Shao X, et al. GLI1 overexpression promotes gastric cancer cell proliferation and migration and induces drug resistance by combining with the AKT-mTOR pathway. Biomed Pharmacother. 2019;111:993–1004. doi: 10.1016/j.biopha.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 80.Ertao Z, Jianhui C, Chuangqi C, Changjiang Q, Sile C, Yulong H, Hui W, Shirong C. Autocrine Sonic hedgehog signaling promotes gastric cancer proliferation through induction of phospholipase Cgamma1 and the ERK1/2 pathway. J Exp Clin Cancer Res. 2016;35:63. doi: 10.1186/s13046-016-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JY, Ko GH, Lee YJ, Ha WS, Choi SK, Jung EJ, et al. Prognostic value of sonic hedgehog protein expression in gastric cancer. Jpn J Clin Oncol. 2012;42(11):1054–1059. doi: 10.1093/jjco/hys137. [DOI] [PubMed] [Google Scholar]
- 82.Yoo YA, Kang MH, Lee HJ, Kim BH, Park JK, Kim HK, Kim JS, Oh SC. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71(22):7061–7070. doi: 10.1158/0008-5472.CAN-11-1338. [DOI] [PubMed] [Google Scholar]
- 83.Niu Y, Li F, Tang B, Shi Y, Hao Y, Yu P. Clinicopathological correlation and prognostic significance of sonic hedgehog protein overexpression in human gastric cancer. Int J Clin Exp Pathol. 2014;7(8):5144–5153. [PMC free article] [PubMed] [Google Scholar]
- 84.Ryu HS, Park DJ, Kim HH, Kim WH, Lee HS. Combination of epithelial–mesenchymal transition and cancer stem cell-like phenotypes has independent prognostic value in gastric cancer. Hum pathol. 2012;43(4):520–528. doi: 10.1016/j.humpath.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 85.Norollahi SE, Mansour-Ghanaei F, Joukar F, Ghadarjani S, Mojtahedi K, Gharaei Nejad K, et al. Therapeutic approach of Cancer stem cells (CSCs) in gastric adenocarcinoma; DNA methyltransferases enzymes in cancer targeted therapy. Biomed Pharmacother. 2019;115:108958. doi: 10.1016/j.biopha.2019.108958. [DOI] [PubMed] [Google Scholar]
- 86.Kumagai K, Tanaka T, Yamagata K, Yokoyama N, Shimizu K. Liver metastasis in gastric cancer with particular reference to lymphatic advancement. Gastric Cancer. 2001;4(3):150–155. doi: 10.1007/PL00011738. [DOI] [PubMed] [Google Scholar]
- 87.Kurihara N, Kubota T, Furukawa T, Watanabe M, Otani Y, Kumai K, Kitajima M. Chemosensitivity testing of primary tumor cells from gastric cancer patients with liver metastasis can identify effective antitumor drugs. Anticancer Res. 1999;19(6B):5155–5158. [PubMed] [Google Scholar]
- 88.Brungs D, Aghmesheh M, Vine KL, Becker TM, Carolan MG, Ranson M. Gastric cancer stem cells: evidence, potential markers, and clinical implications. J Gastroenterol. 2016;51(4):313–326. doi: 10.1007/s00535-015-1125-5. [DOI] [PubMed] [Google Scholar]
- 89.Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, Gordon SA, Shimada Y, Wang TC. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27(5):1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Han SA, Jang JH, Won KY, Lim SJ, Song JY. Prognostic value of putative cancer stem cell markers (CD24, CD44, CD133, and ALDH1) in human papillary thyroid carcinoma. Pathol Res Pract. 2017;213(8):956–963. doi: 10.1016/j.prp.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Kapucuoglu N, Bozkurt KK, Baspinar S, Kocer M, Eroglu HE, Akdeniz R, Akcil M. The clinicopathological and prognostic significance of CD24, CD44, CD133, ALDH1 expressions in invasive ductal carcinoma of the breast: CD44/CD24 expression in breast cancer. Pathol Res Pract. 2015;211(10):740–747. doi: 10.1016/j.prp.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 92.Okudela K, Woo T, Mitsui H, Tajiri M, Masuda M, Ohashi K. Expression of the potential cancer stem cell markers, CD133, CD44, ALDH1, and beta-catenin, in primary lung adenocarcinoma–their prognostic significance. Pathol Int. 2012;62(12):792–801. doi: 10.1111/pin.12019. [DOI] [PubMed] [Google Scholar]
- 93.Padua D, Figueira P, Ribeiro I, Almeida R, Mesquita P. The relevance of transcription factors in gastric and colorectal cancer stem cells identification and rradication. Front Cell Dev Biol. 2020;8:442. doi: 10.3389/fcell.2020.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carrasco-Garcia E, Garcia-Puga M, Arevalo S, Matheu A. Towards precision medicine: linking genetic and cellular heterogeneity in gastric cancer. Ther Adv Med Oncol. 2018;10:8. doi: 10.1177/1758835918794628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Padua D, Barros R, Amaral AL, Mesquita P, Freire AF, Sousa M, et al. A SOX2 reporter system identifies gastric cancer stem-like cells sensitive to Monensin. Cancers (Basel) 2020;12(2):495. doi: 10.3390/cancers12020495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang MC, Li CL, Cui J, Jiao M, Wu T, Jing LI, Nan KJ. BMI-1, a promising therapeutic target for human cancer. Oncol Lett. 2015;10(2):583–588. doi: 10.3892/ol.2015.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, Zhe H, Ding Z, Gao P, Zhang N, Li G. Cancer stem cell marker Bmi-1 expression is associated with basal-like phenotype and poor survival in breast cancer. World J Surg. 2012;36(5):1189–1194. doi: 10.1007/s00268-012-1514-3. [DOI] [PubMed] [Google Scholar]
- 98.Sari IN, Phi LTH, Jun N, Wijaya YT, Lee S, Kwon HY. Hedgehog signaling in cancer: a prospective therapeutic target for eradicating cancer stem cells. Cells. 2018;7(11):208. doi: 10.3390/cells7110208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu X, Su B, Xie C, Wei S, Zhou Y, Liu H, et al. Sonic hedgehog-Gli1 signaling pathway regulates the epithelial mesenchymal transition (EMT) by mediating a new target gene, S100A4, in pancreatic cancer cells. PLoS ONE. 2014;9(7):e96441. doi: 10.1371/journal.pone.0096441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nedjadi T, Salem N, Khayyat D, Al-Sayyad A, Al-Ammari A, Al-Maghrabi J. Sonic hedgehog expression is associated with lymph node invasion in urothelial bladder cancer. Pathol Oncol Res. 2019;25(3):1067–1073. doi: 10.1007/s12253-018-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fang M, Wu JR, Lai X, Ai HY, Tao YF, Zhu B, Huang LS. CD44 and CD44v6 are correlated with gastric cancer progression and poor patient prognosis: evidence from 42 studies. Cell Physiol Biochem. 2016;40(3–4):567–578. doi: 10.1159/000452570. [DOI] [PubMed] [Google Scholar]
- 102.Li K, Guo X, Wang Z, Li X, Bu Y, Bai X, Zheng L, Huang Y. The prognostic roles of ALDH1 isoenzymes in gastric cancer. Onco Targets Ther. 2016;9:3405–3414. doi: 10.2147/OTT.S102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Search strategy and syntax in different databases based on the expression of cancer stem cell markers in gastric cancer.
Data Availability Statement
Not applicable.