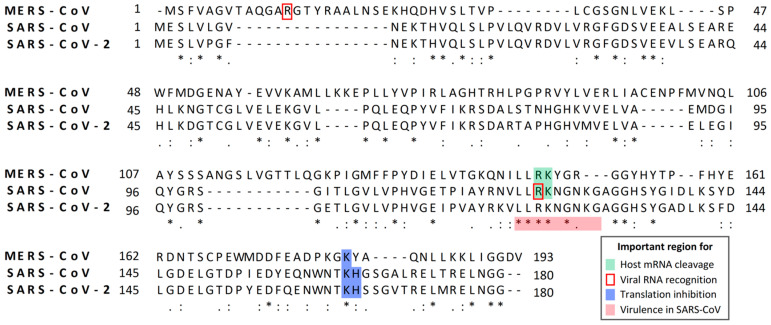
Figure 2.
Alignment of the amino acid sequences of nsp1s of MERS-CoV, SARS-CoV, and SARS-CoV-2. Nsp1 sequences of the MERS-CoV strain EMC2012 (accession no.: YP_009047229), SARS-CoV strain Urbani (accession no.: AAP13442), and SARS-CoV-2 isolate Wuhan-Hu-1 (accession no.: MN908947.3) are aligned using Multiple Sequence Comparison by Log-Expectation (MUSCLE) alignment algorithm. Perfect matches, high-amino acid similarities, and low-amino acid similarities are represented by asterisks, double dots, and single dots, respectively. A dash “-” indicates a gap in the sequence. The numbers beside the aligned sequences show the positions of amino acid residues. Residues shown in green represent the functional amino acids for host mRNA cleavage. Residues shown by a red box outline represent the functional amino acids for viral mRNA recognition. Residues shown in blue represent the amino acids that are important for translation inhibition. Residues shown in red represent the amino acids that are important for virulence in mice.