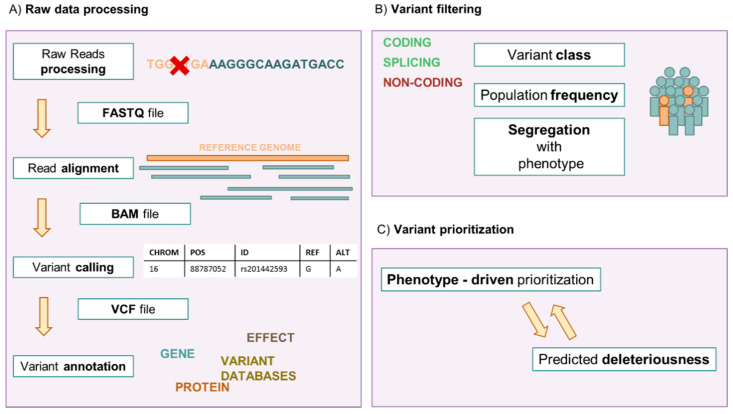
Figure 2.
NGS bioinformatic workflow. (A) The sequencing reaction generates millions of short reads (40 to 400 nucleotides long). The reads are processed by marking duplicates and barcode and adapter sequences. Individual reads are retained in a FASTQ file. They are then aligned to the reference genome, generating a BAM file. Variants are identified (called) from nucleotide positions differing from the reference sequence, and gathered in a VCF file, consisting of a list of genomic coordinates with the reference sequences, the putative variants and quality scores. These variants are then annotated with information gathered from various databases, on variant frequency, gene(s) involved, gene products, predicted deleteriousness, reported pathogenicity or benignity. They are then manually analyzed by filtering (B) and prioritization (C). Variants with no impact on gene products, high frequency in the general population and not segregating with the phenotype are ruled out. The remaining variants are prioritized by various criteria, for example by potential relation to the phenotype or predicted deleteriousness.