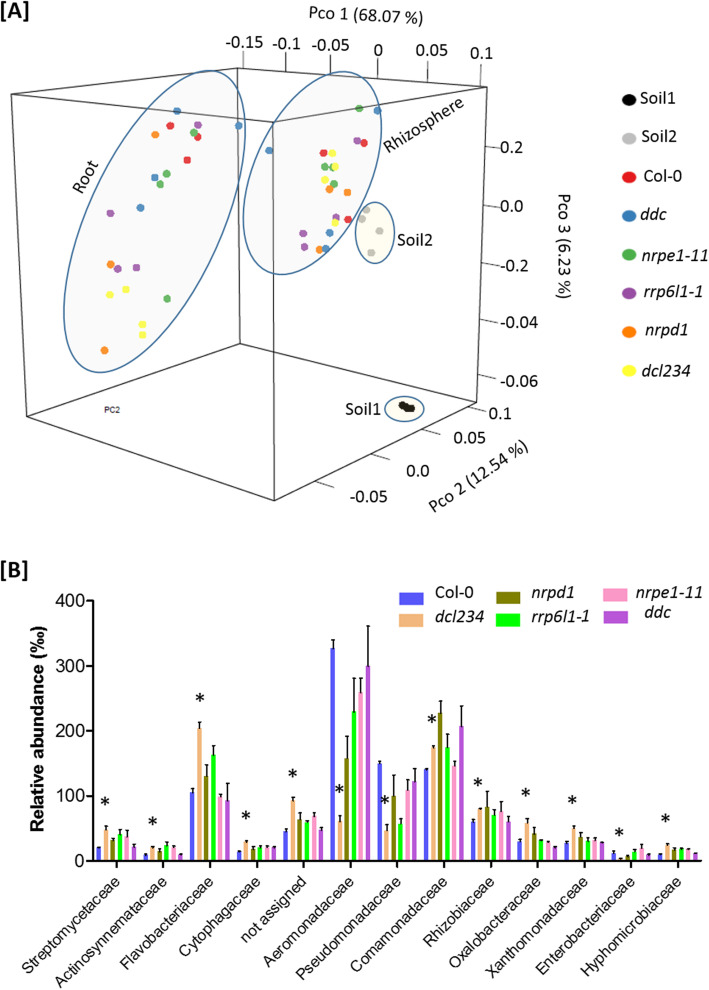
Fig. 1.
Arabidopsis root-associated microbiota is altered in the dcl234 mutant. a PCoA using weighted UniFrac metrics indicates that the largest separation between bacterial communities is spatial proximity to the root (PCo 1). The bacteria communities were investigated by 16S rRNA gene sequencing of the metagenomic DNA. n ≥ 3 biological replicates. b Relative abundance of the top 13 abundant families in roots of the wild type Arabidopsis (Col-0) and the RNA-directed DNA methylation pathway mutants. Mean ± SE, n ≥ 3. Asterisks indicate significant difference (FDR ≤ 0.05) between the mutant and the wild type. Taxa with RA > 5‰ in at least one sample were included in the statistical analysis. The bacteria communities were investigated by 16S rRNA gene sequencing of the metagenomic DNA. See also Figures S1, S2, S3, S4