Abstract
Background
Exosomes play important roles in angiogenesis, drug resistance, and metastasis of colorectal cancer (CRC), but the underlying mechanism has seldom been reported. Herein, our study aimed to reveal an exosomal miRNA-mRNA network involved in CRC by performing bioinformatical analysis.
Methods
The mRNA and miRNA data of colon adenocarcinoma and rectal adenocarcinoma were downloaded from The Cancer Genome Atlas (TCGA) database, and exosomal miRNAs data were downloaded from the GEO dataset GSE39833. The differential expression analysis was performed using “limma” and “edgeR”. Target mRNAs of miRNAs were predicted using FunRich 3.1.3, miRNAtap and multiMiR. The candidate mRNAs and exosomal miRNAs were obtained by intersecting two groups of differentially expressed miRNAs and intersection of the differential expressed mRNAs and the target mRNAs, respectively. Key mRNAs and exosomal miRNAs were identified by the least absolute shrinkage and selection operator regression analysis, and used to construct the exosomal miRNA-mRNA network. The network verified was by receiver operating characteristic curve, GEPIA and LinkedOmics. Functional enrichment analysis was also performed for studied miRNAs and mRNAs.
Results
A total of 6568 differentially expressed mRNAs and 531 differentially expressed miRNAs from TCGA data, and 166 differentially expressed exosomal miRNAs in GSE39833 dataset were identified. Next, 16 key mRNAs and five key exosomal miRNAs were identified from the 5284 candidate mRNAs and 61 candidate exosomal miRNAs, respectively. The exosomal miRNA-mRNA network with high connectivity contained 13 hub mRNAs (CBFB, CDH3, ETV4, FOXQ1, FUT1, GCNT2, GRIN2D, KIAA1549, KRT80, LZTS1, SLC39A10, SPTBN2, and ZSWIM4) and five hub exosomal miRNAs (hsa-miR-126, hsa-miR-139, hsa-miR-141, hsa-miR-29c, and hsa-miR-423). The functional annotation revealed that these hub mRNAs were mainly involved in the regulation of B cell receptor signaling pathway and glycosphingolipid biosynthesis related pathways. All hub mRNAs and hub exosomal miRNAs exhibited high diagnosis value for CRC. Furthermore, the association of the hub mRNAs with overall survival, stages, and MSI phenotype of CRC revealed their important roles in CRC progression.
Conclusion
This study constructed an exosomal miRNA-mRNA network which may play crucial roles in the carcinogenesis and progression of CRC, thus providing potential diagnostic biomarkers and therapeutic targets for CRC.
Keywords: Colorectal cancer, Exosome, Differential expression analysis, Machine learning, MiRNA
Background
As one of the common malignant tumors, colorectal cancer (CRC) is the third leading cause of cancer death worldwide [1, 2]. In recent years, the rapid increases in both CRC incidence and mortality are becoming a heavy burden in many countries [1]. Accurate early diagnosis can enable CRC patients to receive timely and accurate treatment, thereby reducing CRC mortality. It is common to perform a colonoscopy in patients with bowel symptoms due to the suspicion of CRC, but the value of symptoms as indicators of CRC is poor [3]. Some researchers previously proposed that several biomarkers, including carcinoembryonic antigen (CEA) and calprotectin, might be used as a predictor of CRC for symptomatic patients. However, the relatively low specificity and sensitivity of such markers preclude their use for the early diagnosis of CRC [3, 4]. Biopsy plays a decisive role in early cancer and polyposis detection in CRC, which is also important in the differential diagnosis of cancer. This method has made it possible to determine the nature, histological type, and malignancy of the tumor, and can help determine the prognosis, and guide the clinical treatment. However, it is still unpopular in clinical applications because of its cumbersomeness and the difficulty in obtaining a satisfactory specimen [5]. Therefore, it is necessary to identify accurate biomarkers for CRC treatment to facilitate the accurate early diagnosis of grading tumor stage and lesion, and to direct the treatment in CRC [6].
Exosomes are tiny goblet shaped vesicles, 30–140 nm in diameter, that are secreted by cells including immune cells, nerve cells, stem cells, and tumor cells [7–9]. Increasing number of studies elucidated the correlation between exosome production and tumorigenesis. Tumor-derived exosomes participate in the exchange of genetic information between tumor cells and basal cells, which results in angiogenesis and promotes tumor growth and invasion. Recently, exosomes that contain various RNA and protein components have become the focus of intensive research as tumor markers for diagnosis and treatment [10, 11]. Identifying useful biomarkers in exosomes of CRC for diagnosis, prediction of prognosis, and treatment response have achieved great progress in recent years. It has been demonstrated that several exosomal miRNAs, such as miR-125a-3p [12] and miR-638 [13], might be useful as diagnostic biomarkers in CRC. Besides, Ogata-Kawata, Izumiya [14] suggested that seven exosomal miRNAs including miR-223, let-7a, and miR-150 were significantly related to CRC, and most of these miRNAs showed a higher sensitivity for detection of CRC than CEA. All the evidence described above highlights the important role of exosomal miRNA in CRC, as this has become a promising field for finding CRC biomarkers. Although the miRNA-driven mechanisms of cancer have been investigated through many studies, their molecular mechanisms remain unclear and there are still many unidentified exosomal miRNAs associated with tumors. Therefore, further in-depth studies are needed to identify exosomal miRNAs that may serve as potentially effective targets in CRC diagnosis and treatment. Meanwhile, the exploration of regulatory networks between exosomal miRNAs and mRNAs is beneficial for comprehensively understanding the molecular mechanism of CRC development.
Rapid technological advancements in the big data era have spurred the development of bioinformatics into a fast-growing field with applications in a wide range of areas including medical research. Bioinformatics analysis based on high-throughput platforms is an efficient and promising tool to identify biomarkers for diagnosis and prognosis of various cancers [15, 16]. Exploring the interactions of genetic components is essential to understand the mechanisms of carcinogenesis and tumor progression. Scholars previously established and validated a miRNA-mRNA network to reveal the regulatory mechanisms of miRNA in CRC [17]. Besides, Li et al. [18] uncovered a miRNA-regulatory network, and identified two key miRNAs which exhibited dominant regulatory activities in malignant progression of glioma. However, the number of genes exceeds the number of individuals in most datasets, making the interactions discovered by conventional data analytical approaches unreliable. To deal with this issue, one of machine learning approaches, the least absolute shrinkage and selection operator (LASSO) has been increasingly applied to identify the genes correlated with tumor progression and estimate the coefficients in the Cox model [19, 20]. After determining the value of tuning parameter by a cross-validation, the LASSO shrinks most of the coefficients towards zero according to tuning parameter, thereby maximizing the out-of-data prediction accuracy.
In this study, based on TCGA and GSE39833 datasets, differentially expressed exosomal miRNAs were identified by integrating multiple bioinformatics analysis methods. The LASSO approach was subsequently conducted for the identification of key mRNAs and key exosomal miRNAs. An online prediction database, microRNA Data Integration Portal (miRDIP) v4.1 was used to obtain interactions with high confidence class between exosomal key miRNAs and target mRNAs. After importing the interactions into Cytoscape, an exosomal miRNA-mRNA network consisting of 13 mRNAs and five exosomal miRNAs was obtained. The receiver operating characteristic (ROC) curve analysis was applied to validate the diagnostic efficacy of RNAs from the network for CRC. Further analyses uncovered the roles of these 13 mRNAs in CRC progression. Our findings serve as a valuable resource to further explore the mechanisms of CRC development and progression, and provide potentially effective diagnostic markers and therapeutic targets for CRC.
Methods
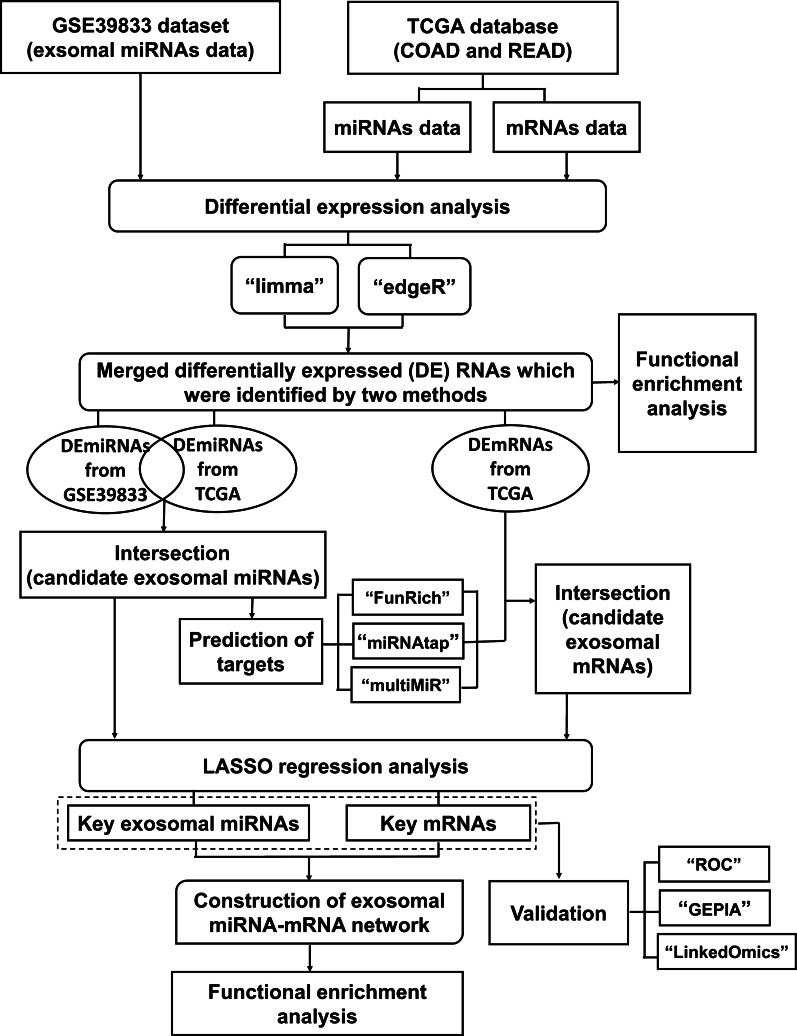
The workflow of this study was described in Fig. 1.
Fig. 1.
The flowchart of this study
Data collection
CRC exosomal miRNA dataset GSE39833 was acquired from the GEO database (https://www.ncbi.nlm.nih.gov/geo/); exosomal miRNAs were collected from the sera of 88 colon cancer patients and 11 healthy controls [14]. The dataset contained 15,739 probe IDs. The mRNA and miRNA data of colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) datasets were downloaded from the cancer genome database (TCGA) (https://portal.gdc.cancer.gov). The TCGA analyze_Normalization function from R package “TCGAbiolinks” was used to filter out extremely lowly expressed genes in TCGA RNA-sequencing data.
Differential expression analysis (DEA)
Using R-packages limma and edgeR [21], we performed the DEA with threshold parameters defined as |log2FC| > 1 and FDR < 0.05 (FC: fold change, FDR: false discovery rate) to identify the differentially expressed mRNAs and miRNAs related to both COAD and READ in TCGA database. In the meanwhile, the differentially expressed exosomal miRNAs were identified based on the GSE39833 dataset. The differentially expressed mRNAs and miRNAs identified by limma and edgeR were visualized using volcano plots, respectively. The results for the two types of DEA methods from the same data were directly merged before conducting subsequent studies.
Prediction of target mRNAs for exosomal miRNAs
The differentially expressed miRNAs identified from TCGA data and GSE39833 dataset were intersected to obtain the candidate exosomal miRNAs that were closely related to CRC. The target mRNAs of the candidate exosomal miRNAs were predicted by using FunRich 3.1.3, and R-packages miRNAtap and multiMiR, respectively. After merging the target mRNAs predicted by these three tools, target mRNAs potentially involved in regulation of CRC progression by exosomal miRNAs (our study defined these mRNAs as candidate mRNAs) were acquired by intersection of the differentially expressed mRNAs and target mRNAs.
Identification of key mRNAs and exosomal miRNAs
As a powerful algorithm, LASSO is used for extracting relevant factors from a large number of unrelated factors. Thus, the LASSO approach was applied to identify the key mRNAs and key exosomal miRNAs that are closely related to CRC from the large number of candidate exosomal miRNAs and candidate mRNAs with the contribution of each RNA being weighted using relative coefficients. The optimal number (λ) of key mRNA and key exosomal miRNAs in combination was determined based on the minimum cross-validated error.
It should be mentioned that, to resolve the data imbalance problem that the number of control samples (n) were obviously less than that of tumor (m), the feature selection was performed multiple times in this study. Each time, n samples were randomly extracted from m tumor samples, and subsequently combined with all of the control samples (n) as the training sets for feature selection. Finally, the times that all features have been screened out in the feature selection were counted, those features with high counts were considered as key features [22]. In this study, LASSO algorithm was performed as the feature selection method, and ten-fold cross validation was used to evaluate the robustness of the feature and parameter λ. The selections of candidate exosomal miRNAs and candidate mRNAs were performed a thousand and ten thousand times, respectively. The candidate exosomal miRNAs and mRNAs were considered as the key exosomal miRNAs and their target mRNAs, respectively, when counts of their occurrences in LASSO regression results were more than 500 and 5000.
Exosomal miRNA-mRNA network model construction
By using miRDIP v4.1 online tool, the interactions between the key mRNAs and key exosomal miRNAs were obtained. Then, the interactions showing high confidence class were chosen for construction of exosomal miRNA-mRNA network by using Cytoscape v3.7.2. The mRNAs in the network were considered as hub mRNAs in this study.
Validation of the hub mRNAs and key exosomal miRNAs
The ROC curve analysis was applied to validate the hub mRNAs and key exosomal miRNAs. GEPIA (http://gepia.cancer-pku.cn/) is a web portal for analyzing cancer type and multiple gene data. It provides easy access to public cancer type and gene data and allows users to use advanced computers to validate biomarkers or potential genes of interest. The overall survival of the key mRNAs was performed using GEPIA. The expression levels of the mRNAs at different stages of CRC were then conducted. Besides, the LinkedOmics (http://www.linkedomics.org/admin.php) was applied to perform the expression levels of the hub mRNAs in microsatellite instability (MSI) phenotype of CRC.
Results
Differential expression analysis of the mRNA and miRNA in COAD and READ
To identify the differentially expressed mRNAs and miRNAs in COAD and READ, and differentially expressed exosomal miRNA between CRC patients and healthy individuals, we performed DEA using the R packages of “limma” and “edgeR”. The results were shown in Table 1. The differentially expressed mRNA and miRNA were visualized by volcano plots. The volcano plots for the result of “limma” were displayed in Figs. 2a, 3a and 4a, while those for the result of “edgeR” were displayed in Figs. 2b, 3b and 4b. After merging the results of “limma” and “edgeR”, a total of 6567 differentially expressed mRNAs, 530 differentially expressed miRNAs, and 165 differentially expressed exosomal miRNAs were obtained. The identified differentially expressed mRNAs and miRNAs were listed in Additional file 1: Table S1.
Table 1.
Results of differentially expressed mRNAs and miRNAs from TCGA, and exosomal miRNAs of GSE39833
| Compare | Style | Identification method | Log2FC_Cutoff | padj_Cutoff | All differential expressed Num | Up regulated Num | Down regulated Num | Total Num after merging |
|---|---|---|---|---|---|---|---|---|
| Tumor versus normal (TCGA-COADREAD) | mRNA | limma | 1 | 0.05 | 5621 | 2585 | 3036 | 6567 |
| edgeR | 1 | 0.05 | 5921 | 2881 | 3040 | |||
| Tumor versus normal (TCGA-COADREAD) | miRNA | limma | 1 | 0.05 | 476 | 161 | 315 | 530 |
| edgeR | 1 | 0.05 | 502 | 251 | 251 | |||
| Tumor versus normal (GSE39833) | Exosomal miRNA | limma | 1 | 0.05 | 79 | 38 | 41 | 165 |
| edgeR | 1 | 0.05 | 274 | 244 | 30 |
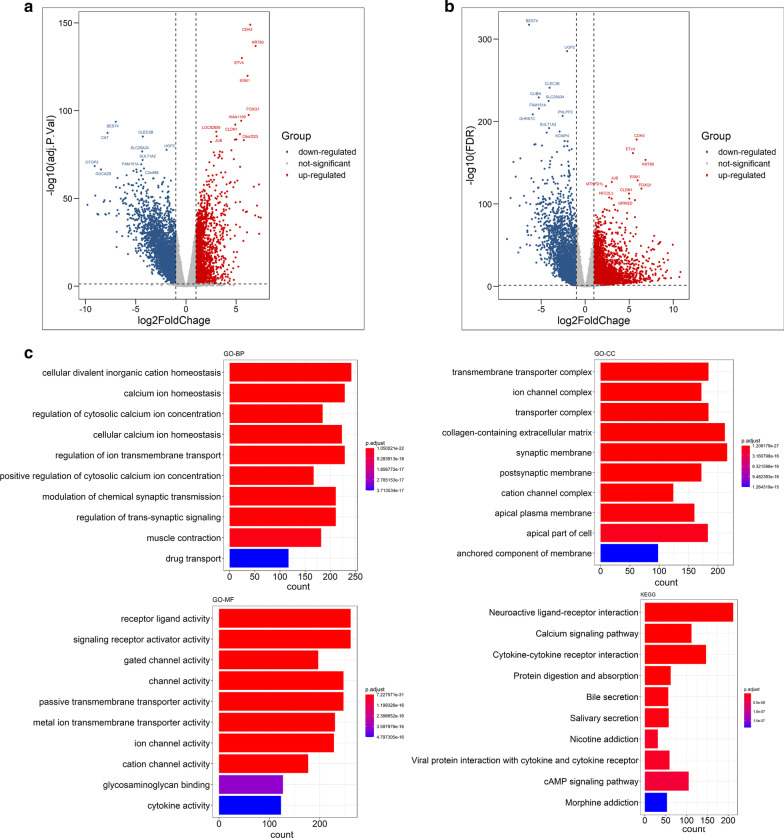
Fig. 2.
Results of differentially expression analysis in mRNAs from TCGA. The volcano plots of the differentially expressed mRNAs identified by a “limma” and b “edgeR”, respectively. c The functional enrichment analysis of differentially expressed mRNAs obtained by merging the results of “limma” and “edgeR”
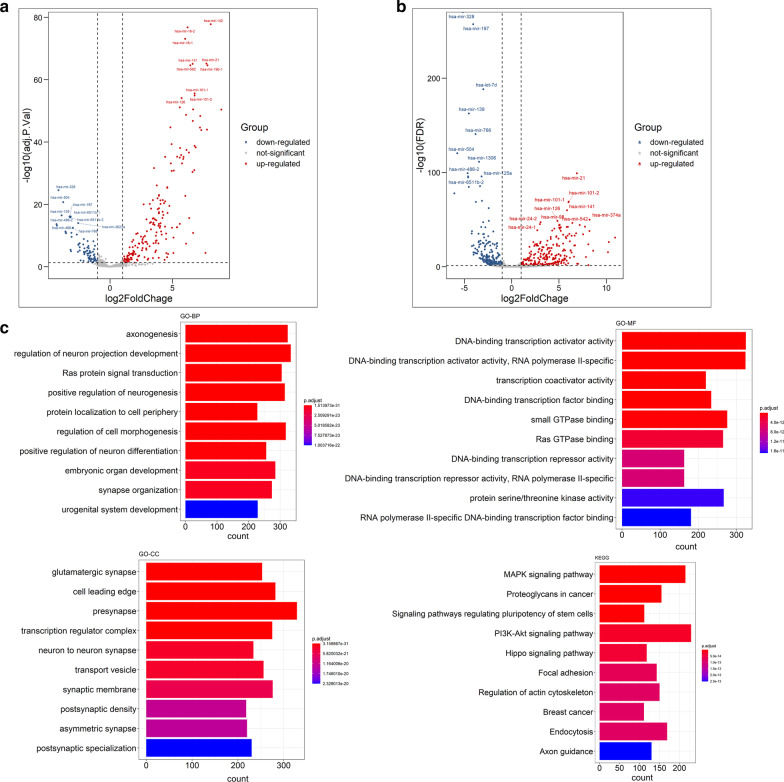
Fig. 3.
Results of differentially expression analysis of miRNAs from TCGA. The volcano plots of the differentially expressed mRNAs identified by a “limma” and b “edgeR”, respectively. c The functional enrichment analysis of differentially expressed miRNAs obtained by merging the results of “limma” and “edgeR”
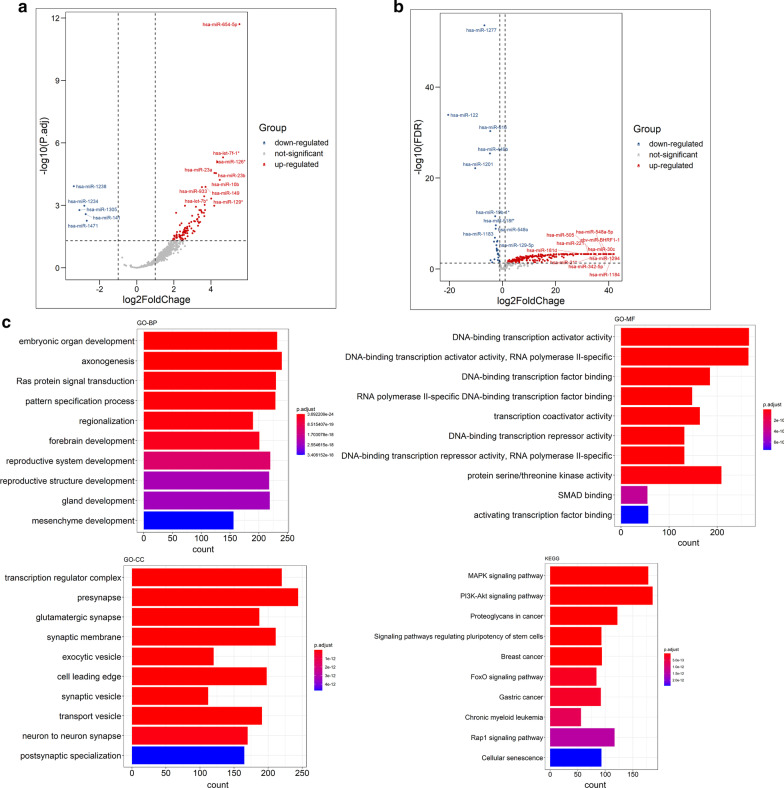
Fig. 4.
Results of differentially expression analysis in exosomal miRNAs from GSE39833 dataset. The volcano plots of the differentially expressed exosomal miRNAs identified by a “limma” and b “edgeR”, respectively. c The functional enrichment analysis of differentially expressed exosomal miRNAs obtained by merging the results of “limma” and “edgeR”
The functional enrichment analysis of the differentially expressed mRNAs revealed that they were mainly involved in processes such as cellular divalent inorganic cation homeostasis and calcium ion homeostasis in biological process (GO-BP), transmembrane transporter/ion channel complex and collagen-containing extracellular matrix in cellular component (GO-CC), and channel activity, passive transmembrane transporter activity and receptor ligand activity in molecular function (GO-MF). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed the interaction of neuroactive ligand-receptor, cytokine-cytokine receptor, and the signaling pathway of calcium and cAMP (Fig. 2c).
The target genes of differentially expressed miRNAs were mainly enriched in processes such as signal transduction of Ras protein, axonogenesis and embryonic organ development in GO-BP; the presynapse, cell leading edge, actin cytoskeleton, and transcription regulator complex in GO-CC; DNA-binding transcription related activities and small/Ras GTPase binding in GO-MF. The KEGG pathway analysis revealed the involvement of genes related to the proteoglycans in cancer, PI3k-Akt and MAPK signaling pathway (Fig. 3c).
As shown in Fig. 4c, the target genes of the differentially expressed exosomal miRNAs were mainly enriched in embryonic organ development and Ras protein signal transduction in GO-BP, presynapse and transcription factor complex in GO-CC, DNA-binding transcription activator activity and DNA-binding transcription activator activity, RNA polymerase II-specific in GO-MF, and proteoglycans in cancer, PI3k-Akt and MAPK signaling pathways in KEGG pathway analysis.
Screening of candidate mRNAs and exosomal miRNAs
To identify exosomal RNAs involved in CRC development, we performed overlap analysis. After merging the differentially expressed exosomal miRNA data (GSE39833) and the differentially expressed miRNAs in TCGA, we found 61 common miRNAs that showed a consensus between the two sets of miRNA data (Table 2). These 61 miRNAs were considered as candidate exosomal miRNAs. After predicting target mRNAs for the 61 candidate exosomal miRNAs, a total of 18,641 target mRNAs were obtained (Additional file 2: Table S2). Finally, a total of 5284 candidate mRNAs were acquired by intersection of these target mRNAs with the differentially expressed mRNAs.
Table 2.
The results of the consistent miRNAs between differentially expressed miRNA and exosomal miRNAs
| MicroRNA symbol | MicroRNA symbol | MicroRNA symbol |
|---|---|---|
| hsa-let-7c | hsa-mir-150 | hsa-mir-337 |
| hsa-let-7d | hsa-mir-1539 | hsa-mir-362 |
| hsa-mir-100 | hsa-mir-181d | hsa-mir-421 |
| hsa-mir-106b | hsa-mir-192 | hsa-mir-423 |
| hsa-mir-10a | hsa-mir-193b | hsa-mir-424 |
| hsa-mir-10b | hsa-mir-197 | hsa-mir-484 |
| hsa-mir-1180 | hsa-mir-206 | hsa-mir-495 |
| hsa-mir-1224 | hsa-mir-22 | hsa-mir-539 |
| hsa-mir-1227 | hsa-mir-221 | hsa-mir-545 |
| hsa-mir-1228 | hsa-mir-224 | hsa-mir-552 |
| hsa-mir-1229 | hsa-mir-23a | hsa-mir-574 |
| hsa-mir-1234 | hsa-mir-23b | hsa-mir-605 |
| hsa-mir-1237 | hsa-mir-27a | hsa-mir-654 |
| hsa-mir-1238 | hsa-mir-27b | hsa-mir-671 |
| hsa-mir-126 | hsa-mir-296 | hsa-mir-760 |
| hsa-mir-1306 | hsa-mir-29a | hsa-mir-766 |
| hsa-mir-1307 | hsa-mir-29c | hsa-mir-770 |
| hsa-mir-130a | hsa-mir-301a | hsa-mir-92b |
| hsa-mir-139 | hsa-mir-30a | hsa-mir-95 |
| hsa-mir-141 | hsa-mir-30b | |
| hsa-mir-149 | hsa-mir-328 |
There were 61 efficient miRNAs in exosomes
Construction of exosomal miRNA-mRNA network
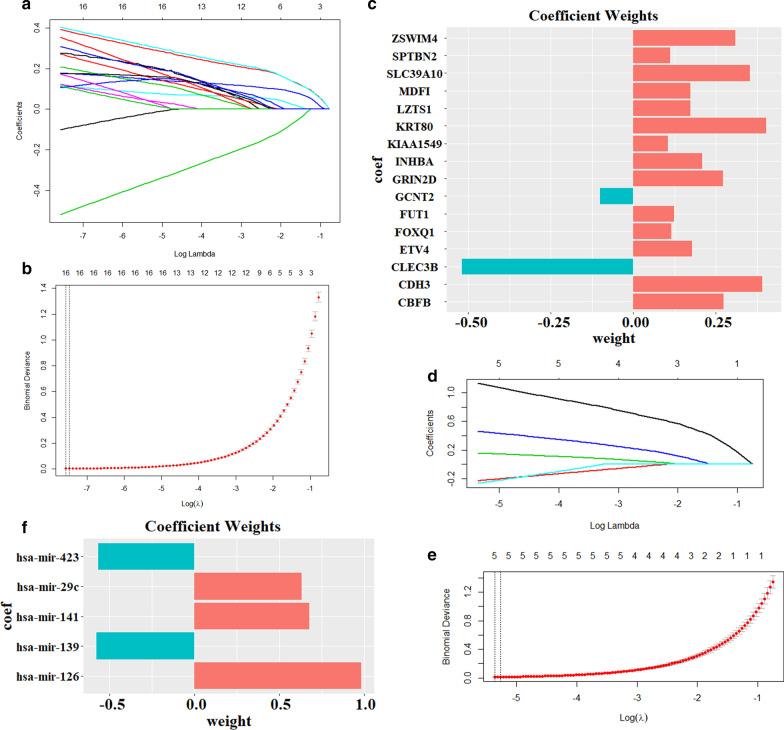
The path coefficient and cross-validation binomial deviance curve for logistic LASSO were carried out to minimize overfitting in the models of mRNAs or miRNAs, and the optimal lambda of mRNA model (λ = 16) and miRNA model (λ = 5) were subsequently obtained (Fig. 5a, b, d, e). A total of 16 key mRNAs and five key exosomal miRNAs were identified, and their coefficient weights were depicted in Fig. 5c, f, respectively. By using the bidirectional search function in the mirDIP database, 25 bidirectional relationships with high confidence class were obtained after entering the key mRNAs and exosomal miRNAs.
Fig. 5.
Identification of key mRNAs and exosomal miRNAs by LASSO regression approach. a Tuning parameter of LASSO model selection, b LASSO coefficient profiles, and c coefficient values for the 16 key mRNAs. d Tuning parameter of LASSO model selection, e LASSO coefficient profiles, and f coefficient values for the five key exosomal miRNAs
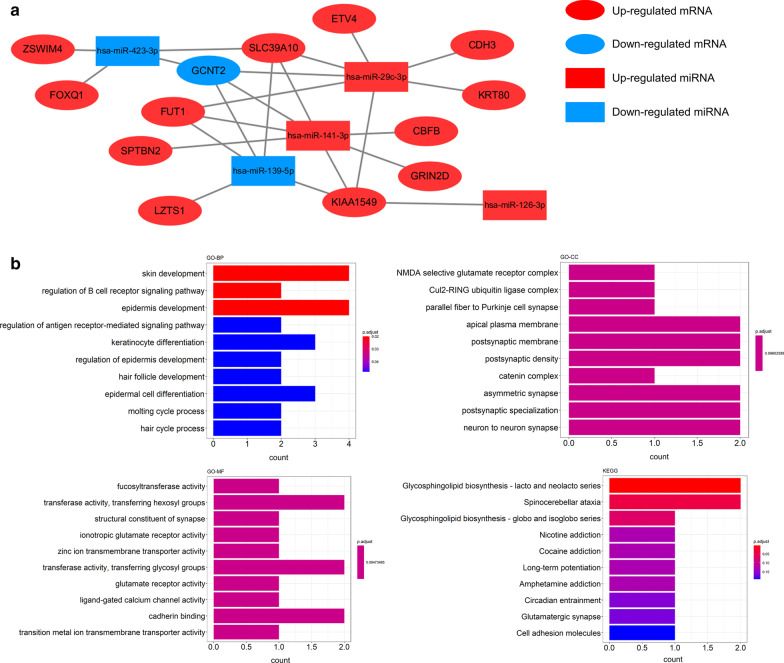
As shown in Fig. 6a, an exosomal miRNA-mRNA network consisted of five miRNAs (hsa-miR-126, hsa-miR-139, hsa-miR-141, hsa-miR-29c, and hsa-miR-423) and 13 mRNAs (CBFB, CDH3, ETV4, FOXQ1, FUT1, GCNT2, GRIN2D, KIAA1549, KRT80, LZTS1, SLC39A10, SPTBN2, and ZSWIM4) was constructed. These mRNAs and miRNAs were respectively considered to be hub mRNAs and hub exosomal miRNAs that might play crucial roles in CRC development via exosomes. According to the network, we found that the hsa-miR-141 and hsa-miR-29c interacted with more than half of mRNAs. Meanwhile, FUT1, GCNT2, KIAA1549 and SLC39A10 were connected with four of five exosomal hub miRNAs. These findings suggested hsa-miR-141, hsa-miR-29c, FUT1, GCNT2, KIAA1549 and SLC39A10 might be key contributors in the development of CRC. Additionally, the interactions of these miRNA/mRNA also might provide novel insights into the molecular mechanism of the roles of exosomes in CRC. As shown in Fig. 6b, the functional annotation of the 13 hub mRNAs showed that they were mainly enriched in skin development and regulation of B cell receptor signaling pathway in GO-BP, apical plasma membrane and postsynaptic membrane in GO-CC, transferase activity, transferring hexosyl/glycosyl groups in GO-MF, and glycosphingolipid biosynthesis related signaling pathways and Nicotine/Cocaine addiction in the KEGG pathway analysis.
Fig. 6.
Analysis of hub mRNAs/exosomal miRNAs. a Exosomal miRNA-mRNA network. b Functional enrichment analysis of the hub mRNAs
Validation of the hub mRNAs and hub exosomal miRNAs
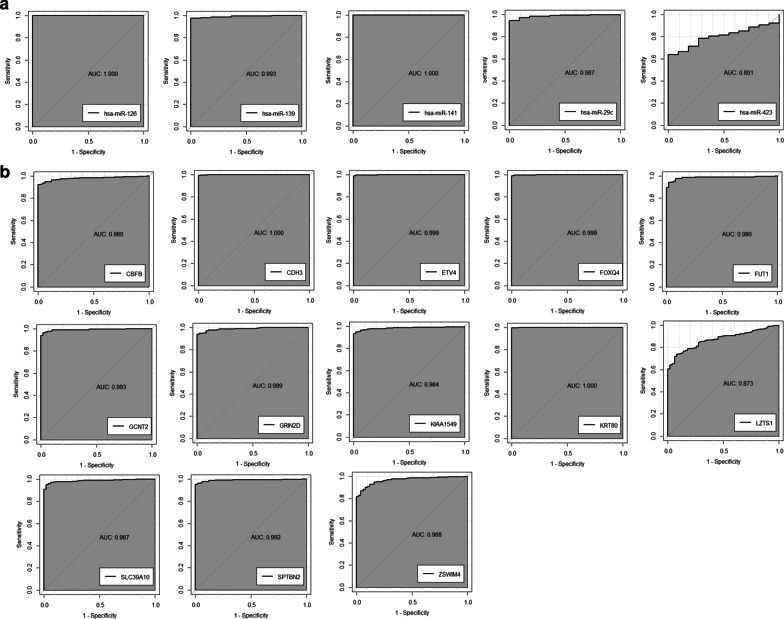
We explored the value of the hub mRNAs and hub exosomal miRNAs as diagnosis biomarkers in CRC by performing ROC curves and calculating the area under the curves (AUCs) [95% confidence intervals (CIs)]. As shown in Fig. 7a, the AUCs of hsa-miR-126, hsa-miR-139, hsa-miR-141, hsa-miR-29c, and hsa-miR-423 were respectively 1.000, 0.993, 1.000, 0.987 and 0.801, which proved that the five hub exosomal miRNAs can well distinguish tumor and normal samples. As shown in Fig. 7b, the AUCs of CBFB, CDH3, ETV4, FOXQ1, FUT1, GCNT2, GRIN2D, KIAA1549, KRT80, LZTS1, SLC39A10, SPTBN2, and ZSWIM4 were respectively 0.980, 1.000, 0.999, 0.999, 0.986, 0.993, 0.989, 0.984, 1.000, 0.873, 0.987, 0.992, 0.968, and 0.904, suggesting that these hub mRNAs had highly diagnostic accuracy potential to distinguish tumors from normal tissues.
Fig. 7.
ROC curve analysis of the a hub mRNAs and b exosomal hub miRNAs
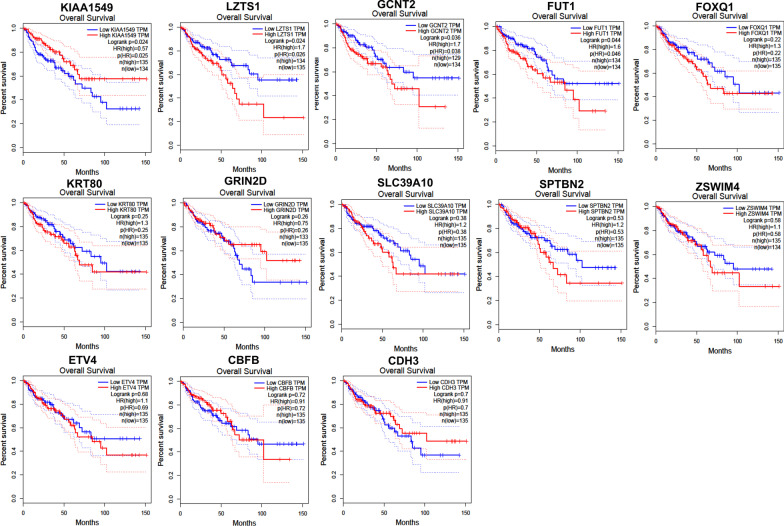
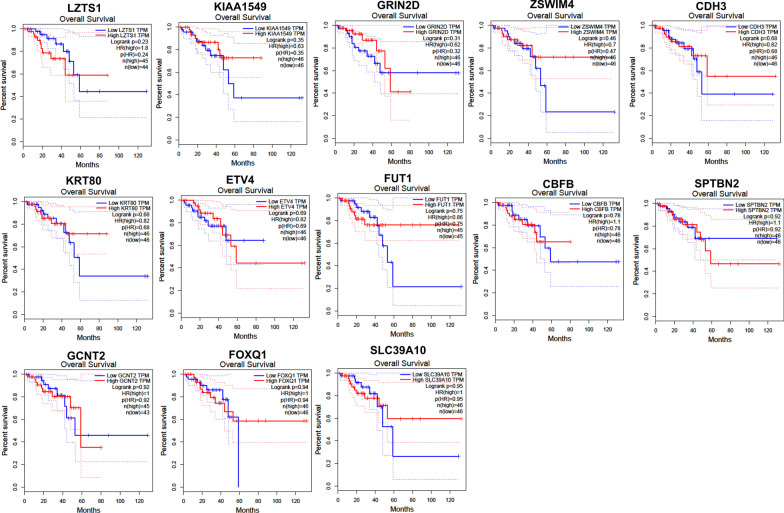
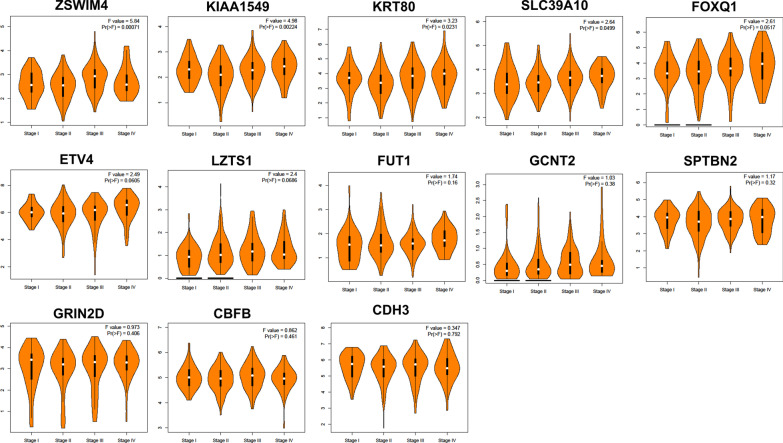
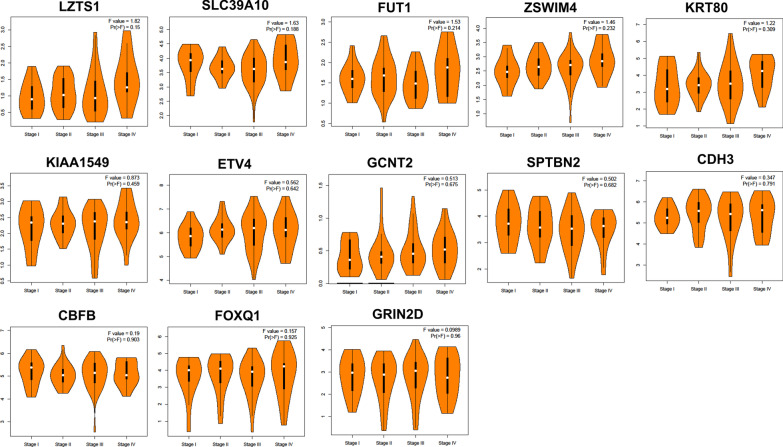
Additionally, we used GEPIA to conduct the survival and stage analyses of the hub mRNAs. The results showed that the expressions of FUT1, GCNT2, KIAA1549 and LZTS1 were significantly related to the overall survival (OS) in COAD (P < 0.05, Fig. 8). All the hub mRNAs were not related to the OS in READ (P > 0.05, Fig. 9). Besides, we found a strong correlation of the COAD stages with expressions of KIAA1549, KRT80, SLC39A10 and ZSWIM4 (P < 0.05, Fig. 10), hinting that these four hub mRNAs might be involved in the COAD stages. Notably, the stage analysis of the hub mRNAs in READ showed a similar result with the survival analysis in READ (Fig. 11). We speculated that it is because the sample size of READ was small in the TCGA database.
Fig. 8.
Results for the overall survival (OS) analysis of the hub mRNAs in COAD (Kaplan–Meier Plotter). Note The dotted lines on the Kaplan–Meier curves represented 95% confidence intervals
Fig. 9.
Results for the overall survival (OS) analysis of the hub mRNAs in READ (Kaplan–Meier Plotter). Note The dotted lines on the Kaplan–Meier curves represented 95% confidence intervals
Fig. 10.
The expression of the hub mRNAs in different tumor stage of COAD patients
Fig. 11.
The expression of the hub mRNAs in different tumor stage of READ patients
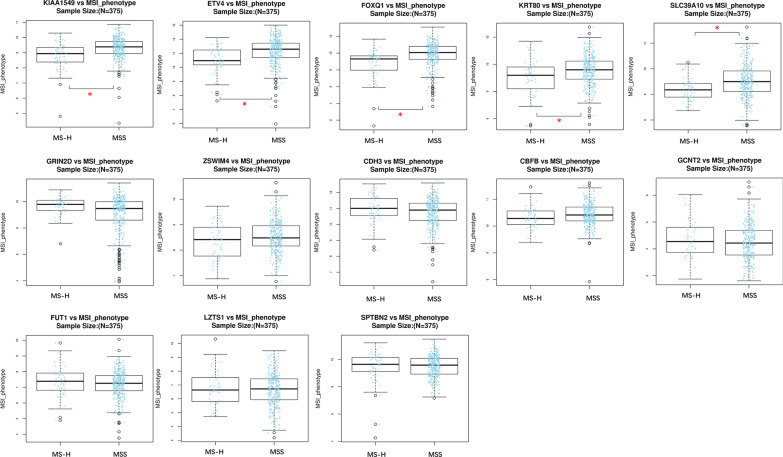
Furthermore, we investigated the relation of these exosomal miRNA-regulated hub mRNAs with the MSI phenotype in CRC. As shown in Fig. 12, the expression levels of ETV4, FOXQ1, KIAA1549, KRT80, and SLC39A10 were significantly different in microsatellite stable (MSS) and microsatellite instability-high (MS-H) subtype CRC.
Fig. 12.
The expression of the hub mRNAs in different MSI phenotype of CRC patients. The expression of ETV4, FOXQ1, KIAA1549, KRT80, and SLC39A10 were significant in distinguishing MS-H and MSS phenotype (P value < 0.05)
Discussion
CRC, including colon and rectal cancer, has become one of the leading causes of cancer in most countries and regions worldwide in the recent decades [23, 24]. Given the high mortality rate of CRC, there is an urgent need for effective prevention and treatment modalities [25]. With the recent advances in the fields of medicine and biotechnology, precise treatment of CRC through gene targeted therapy is a new and efficient potential therapeutic approach for CRC, which requires the identification of marker genes associated with cancer [26]. Multiple studies have proven that exosomes play an important role in cancer development including angiogenesis, drug resistance, and cancer metastasis [27–29]. Therefore, the properties of exosomes are being exploited for diagnostic, prognostic, and therapeutic applications. It was reported that exosome biomarkers can be used for early diagnosis and prognosis of breast cancer [30]. A previous study conducted by Hu and his colleagues demonstrated that exosomal miR-92a-3p was significantly associated with metastasis and chemotherapy resistance in CRC, which may be used for the prediction of prognosis in CRC [31]. However, the underlying mechanism of exosome-regulated CRC progression has not been elucidated. In this study, we aimed to reveal an exosomal miRNA-mRNA network that drives the mechanisms of CRC, and identify potential biomarkers for improving the diagnosis accuracy of CRC.
“limma” and “edgeR” are two approaches of DEA relying on different assumptions and techniques, so the results of the two methods are commonly different. There is no standard to judge which methods obtains more credible results. In order to avoid the possibility of excluding some important RNAs due to only one method, this study used both methods to perform DEA and combine the results for subsequent experiments. Through comprehensive analysis of the open access miRNA and mRNA data for CRC from TCGA, we identified 6567 mRNAs and 530 miRNAs that were differentially expressed in COAD and READ. We also identified 165 differentially expressed exosomal miRNAs based on GSE39833 dataset. It is worth mentioning that the differentially expressed miRNAs and exosomal miRNAs we identified were both mainly involved in MAPK and PI3k-Akt signaling pathways. The MAPK signaling pathway functions as a molecular switch for multiple cellular responses during both homeostasis and pathological conditions [32]. Both the MAPK and PI3k-Akt signaling pathways play a critical regulatory role in the development and progression of CRC [33]. These data indicate that the screened exosomal miRNAs were robust and can potentially affect the tumor fate.
With the aim of screening more robust and potential miRNAs that modulated CRC development via exosomes, we performed intersection of differentially expressed miRNAs with differentially expressed exosomal miRNAs for further studies. After finishing the intersection, a total of 61 candidate exosomal miRNAs were obtained to predict their targeted mRNA. Next, 5284 mRNAs were obtained by overlapping the target mRNAs and differentially expressed mRNAs, which could ensure to get the most potential mRNAs associated with both CRC progression and exosomes. Recently, machine learning approaches have gained popularity in medicine [34, 35]. The prowess and flexibility of machine learning methods enable researchers to extract valuable information from increasing biomedical databases. By integrating a logistic regression model and the LASSO method, scholars established a scoring system that could efficiently predict the risk of thyroid malignancy [36]. A previous study also proved that machine-learning methods might be a prior choice in the analysis of high dimensional data when no prior knowledge is available [37]. Based on the candidate mRNAs and exosomal miRNAs, LASSO method identified five key exosomal miRNAs and 16 key mRNAs, which were significantly weighted. By adopting a LASSO regression analysis, we could efficiently identify the regulators closely associated with CRC progression. Then, we found that all the key exosomal miRNAs we identified were reported in previous studies on cancer. Several studies revealed that the hsa-miR-126 could be used as a biomarker in different cancers, such as non-small-cell lung cancer [38] and pancreatic adenocarcinoma [39]. Besides, hsa-miR-126 has been proven to be a tumor suppressor that can inhibit the growth of NSCLC cells and enhance the cytotoxicity of targeted agents [40]. Multiple studies indicated the crucial roles of hsa-miR-139 in CRC progression [41, 42]. Several researches demonstrated that hsa-miR-139 exhibits tumor-suppressive function by regulating NOTCH-1 in CRC [43, 44]. Notably, in the last year, Zhao et al. [45] successfully constructed a nanoparticle loaded with hsa-miR-139 which showed tumor targeting activity and antitumor ability in CRC model. Some previous studies revealed that hsa-miR-141 is closely related to cancer metastasis. For example, in 2015, Li et al. [46] found that the elevated levels of serum exosomal hsa-miR-141 were significantly correlated with metastasis of prostate cancer. Besides, Yan et al. [47] demonstrated that hsa-miR-141 plays a certain role in colon cancer metastasis. Additionally, Li et al. [48] indicated overexpression of hsa-miR-423 could promote CRC cell proliferation, pointing out a critical role of hsa-miR-423 on CRC development. All these five exosomal miRNAs exhibited high AUCs in ROC curve analysis, suggesting that they could be used as diagnostic markers for CRC.
As known, exosomes contain a variety of small molecules of RNAs, such as miRNAs and long non-coding RNAs, which could be delivered into the target cells to play certain functions. It is documented that these exosomes of RNAs could participate in complex cell-to-cell communications by changing the gene expression in recipient cells [49, 50]. In multiple diseases, serum exosomes transfer miRNAs into the cell of diseased tissue to modulate the disease development. Thus, we used the mRNA from CRC tissue and miRNAs from serum exosomes to establish an exosomal miRNA-mRNA network for understanding the molecular mechanism of exosomal miRNAs in intercellular communications under CRC microenvironment. Afterward, a total of 13 hub mRNAs regulated by exosomal miRNAs were identified for further analyses and validation. Functional enrichment analysis of these mRNAs revealed they were mainly involved in the terms of skin development and glycosphingolipid biosynthesis related signaling pathways. As the typical components of animal cell membranes, glycosphingolipids play key roles in cells proliferation, adhesion, motility and differentiation and its abnormal biosynthesis is usually found in various cancers including CRC [51, 52]. Therefore, we speculated that the hub exosomal miRNAs might change the expression of hub mRNAs to regulate glycosphingolipid biosynthesis, thereby affecting proliferation, migration and invasion of CRC cells. Then, we found that FOXQ1, GCNT2 and KIAA1549 showed the highest interactions (four) with exosomal miRNAs in the network and showed a strong correlation with the survival times of COAD patients. Nakamura et al. [53] revealed that GCNT2 is closely related to CRC metastasis. FOXQ1 has been identified as a tumor promoter of CRC in a study conducted by Liu et al. [54]. Cumulative studies indicated that FOXQ1 is capable of promoting metastasis in diverse cancers [55–57], especially in CRC [54, 58]. Nevertheless, in the current study, the expression of KIAA1549 was not only significantly associated with OS of COAD patients, but also strongly related to stages and MSI phenotypes in CRC. There is no report that explores the role of KIAA1549 in cancers. Therefore, we speculated that KIAA1549 might play a critical role in the mechanism of CRC, and could be a novel and robust target in CRC therapy. SLC39A10, another hub mRNA connected with the four exosomal hub miRNAs, encodes zinc transporter ZIP10 which is involved in cell migration during tumor progression [59]. In this study, we found its expression was significantly different in stages of COAD as well as MSI phenotypes of CRC. Apart from KIAA1549 and GCNT2, we found the expression of FUT1 was also significantly related to survival times of COAD patients. FUT1 was considered as a potential tumor suppressor, its abnormal expression was frequently found in various cancers [60, 61]. Zhou et al. [62] demonstrated that LZTS1 could suppress cell proliferation and prohibit tumor growth in CRC by regulating AKT-mTOR signaling pathway. Besides, both the expression level of KRT80 and FOXQ1 could efficiently distinguish the phenotype of MSS from MS-H in CRC. The MSI status could alter the tumor microenvironment of CRC patients in different ways, thereby influencing the efficacy of immune checkpoint inhibitors in CRC patients. Several studies revealed that CRC patients with MS-H phenotype were more sensitive to immune checkpoint inhibitors than those with MSS phenotype [63, 64]; nevertheless, the underlying mechanism is still obscure. Our study implied that SLC39A10, KRT80, FOXQ1 and KIAA1549 might be involved in the process of MSI and used to be potential biomarkers of MSI phenotype testing. However, we found that all the identified mRNAs that were regulated by exosomal miRNAs showed no significant correlation with prognosis and stages in READ patients, which is possibly due to the small sample size of READ patients. Furthermore, the ROC analysis revealed that the identified key exosomal miRNAs and hub mRNAs are likely to be good biomarkers with high sensitivity and specificity for diagnosis of CRC in patients. Thus, the detection of the hsa-miR-126, hsa-miR-139, hsa-miR-141, hsa-miR-29c, and hsa-miR-423 in exosomes from patient blood may serve as a non-invasive diagnosis biomarkers for CRC.
Based on our results and above-mentioned literature, we suppose that there are multiple regulatory axes related to CRC development in intracellular communications mediated by serum exosomes. The current study suggested that hsa-miR-423, hsa-miR-139, hsa-miR-141, and hsa-miR-29c in serum exosomes can be protected from RNase degradation and were transferred into target cells and subsequently regulated the expression of certain mRNAs in recipient cells. Specifically, both exosomal hsa-miR-141 and hsa-miR-29c may regulate the expression of GCNT2 to modulate CRC metastasis. Exosomal has-miR-423 probably involves in the process of MSI by regulating SLC39A10 and FOXQ1. Moreover, exosomal hsa-miR-139 could target SLC39A10, KIAA1549, FUTI, and LZTS1 to exert several biological functions in CRC development. Nevertheless, the validation of these exosomal miRNA-mRNA pairs is required for elucidating their precise mechanism underlying CRC development.
Collectively, the current study provides a picture of the patho-mechanism of CRC and provides avenues for further investigation and screening of efficient biomarkers for therapeutic intervention in CRC. Some limitations of this study should be noted. First, the normal sample size was relatively small, but we applied random sampling and multiple trials to minimize the deviations. Besides, our finding is a preliminary step to reveal a novel exosomal miRNA-mRNA network for CRC; further experimental verification and large cohorts are required to determine clinical usefulness of the five exosomal hub miRNAs and their target mRNAs as biomarkers for CRC. The diagnostic value of the five hub exosomal miRNAs should be verified in large-scale and multi-center studies of CRC patients, and their potential clinical significance should be evaluated by comparing clinical and pathological characteristics in these samples. The exosomal miRNA-mRNA network should be verified by in vitro and in vivo studies.
Conclusions
Our study efficiently identified several candidate targets (CBFB, CDH3, ETV4, FOXQ1, FUT1, GCNT2, GRIN2D, KIAA1549, KRT80, LZTS1, SLC39A10, SPTBN2, ZSWIM4, hsa-miR-126, hsa-miR-139, hsa-miR-141, hsa-miR-29c, and hsa-miR-423) that can potentially serve as biomarkers in the diagnosis of CRC, and revealed an exosomal miRNA-mRNA network in CRC progression. These findings provide a new direction for diagnosis and treatment of CRC.
Supplementary Information
Additional file 1. Table S1. The differential expressed miRNA/mRNA identified by edgeR and limma.
Additional file 2. Table S2. The targeted mRNA of the candidate exosomal miRNAs.
Acknowledgements
Not applicable.
Abbreviations
- CRC
Colorectal cancer
- CEA
Carcinoembryonic antigen
- LASSO
Least absolute shrinkage and selection operator
- COAD
Colon adenocarcinoma
- ROC
Receiver operating characteristic
- TCGA
The cancer genome database
- READ
Rectal adenocarcinoma
- GEO
Gene Expression Omnibus
- DEA
Differential expression analysis
- BP
Biological process(s)
- GO
Gene Ontology
- CC
Cellular component
- MF
Molecular function
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
Overall survival
Authors' contributions
JM, PW analyzed and interpreted data. PW, JM, LH and JQ collect data and performed WGCNA analysis. JM and PW were major contributors in the manuscript writing. JL, JM and PW designed the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current study are available from NCBI Gene Expression Omnibus (GEO: GSE39833) https://www.ncbi.nlm.nih.gov/geo/ and the cancer genome database (TCGA: COAD, READ) https://portal.gdc.cancer.gov.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jun Ma and Peilong Wang contributed equally to this work
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-021-00905-2.
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Vega P, Valentín F, Cubiella J. Colorectal cancer diagnosis: pitfalls and opportunities. World J Gastrointest Oncol. 2015;7(12):422–433. doi: 10.4251/wjgo.v7.i12.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Roon AC, Karamountzos L, Purkayastha S, Reese GE, Darzi AW, Teare JP, et al. Diagnostic precision of fecal calprotectin for inflammatory bowel disease and colorectal malignancy. Am J Gastroenterol. 2007;102(4):803–813. doi: 10.1111/j.1572-0241.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- 5.Spronk I, Meijers MC, Heins MJ, Francke AL, Elwyn G, van Lindert A, et al. Availability and effectiveness of decision aids for supporting shared decision making in patients with advanced colorectal and lung cancer: results from a systematic review. Eur J Cancer Care (Engl) 2019;28(3):e13079. doi: 10.1111/ecc.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makimoto S, Takami T, Hatano K, Kataoka N, Yamaguchi T, Tomita M, et al. Additional surgery after endoscopic submucosal dissection for colorectal cancer: a review of 53 cases. Int J Colorectal Dis. 2019;34(10):1723–1729. doi: 10.1007/s00384-019-03370-7. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Corbett AL, Taatizadeh E, Tasnim N, Little JP, Garnis C, et al. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019;3(1):011503. doi: 10.1063/1.5087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75(2):193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sedgwick AE, D'Souza-Schorey C. The biology of extracellular microvesicles. Traffic. 2018;19(5):319–327. doi: 10.1111/tra.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannafon BN, Ding W-Q. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14(7):14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, et al. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7(1):4150. doi: 10.1038/s41598-017-04386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan S, Dang G, Zhang X, Jin C, Qin L, Wang Y, et al. Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget. 2017;8(42):72220–72226. doi: 10.18632/oncotarget.19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao C, Zhou C, Zhuang J, Liu L, Liu C, Li H, et al. MicroRNA expression in cervical cancer: novel diagnostic and prognostic biomarkers. J Cell Biochem. 2018;119(8):7080–7090. doi: 10.1002/jcb.27029. [DOI] [PubMed] [Google Scholar]
- 16.Han Z, Li Y, Zhang J, Guo C, Li Q, Zhang X, et al. Tumor-derived circulating exosomal miR-342-5p and miR-574-5p as promising diagnostic biomarkers for early-stage Lung Adenocarcinoma. Int J Med Sci. 2020;17(10):1428–1438. doi: 10.7150/ijms.43500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu J, Tang W, Du P, Wang G, Chen W, Li J, et al. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol. 2012;6:68. doi: 10.1186/1752-0509-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z, et al. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucl Acids Res. 2013;41(22):e203. doi: 10.1093/nar/gkt1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko S, Hirakawa A, Hamada C. Enhancing the lasso approach for developing a survival prediction model based on gene expression data. Comput Math Methods Med. 2015;2015:259474. doi: 10.1155/2015/259474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadaleta F, Van Steen K. Discovering main genetic interactions with LABNet LAsso-based network inference. PLoS ONE. 2014;9(11):e110451. doi: 10.1371/journal.pone.0110451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan K, Yang Y, Zhang Y, Zhao W, Liao L. Normalization method utilizing endogenous proteins for quantitative proteomics. J Am Soc Mass Spectrom. 2020;31(7):1380–1388. doi: 10.1021/jasms.0c00012. [DOI] [PubMed] [Google Scholar]
- 23.Fuccio L, Repici A, Hassan C, Ponchon T, Bhandari P, Jover R, et al. Why attempt en bloc resection of non-pedunculated colorectal adenomas? A systematic review of the prevalence of superficial submucosal invasive cancer after endoscopic submucosal dissection. Gut. 2018;67(8):1464–1474. doi: 10.1136/gutjnl-2017-315103. [DOI] [PubMed] [Google Scholar]
- 24.Røed Skårderud M, Polk A, Kjeldgaard Vistisen K, Larsen FO, Nielsen DL. Efficacy and safety of regorafenib in the treatment of metastatic colorectal cancer: a systematic review. Cancer Treat Rev. 2018;62:61–73. doi: 10.1016/j.ctrv.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Chen N, Li W, Huang K, Yang W, Huang L, Cong T, et al. Increased platelet-lymphocyte ratio closely relates to inferior clinical features and worse long-term survival in both resected and metastatic colorectal cancer: an updated systematic review and meta-analysis of 24 studies. Oncotarget. 2017;8(19):32356–32369. doi: 10.18632/oncotarget.16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149(5):1204–25.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. 2019;1871(2):455–468. doi: 10.1016/j.bbcan.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundararajan V, Sarkar FH, Ramasamy TS. The multifaceted role of exosomes in cancer progression: diagnostic and therapeutic implications [corrected] Cell Oncol (Dordr) 2018;41(3):223–252. doi: 10.1007/s13402-018-0378-4. [DOI] [PubMed] [Google Scholar]
- 29.Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18(1):32. doi: 10.1186/s12943-019-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Ji S, Shao G, Zhang J, Zhao K, Wang Z, et al. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clin Transl Oncol. 2018;20(7):906–911. doi: 10.1007/s12094-017-1805-0. [DOI] [PubMed] [Google Scholar]
- 31.Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18(1):91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L, et al. A Comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers. 2019;11(10):1618. doi: 10.3390/cancers11101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan ML, Wang Y, Zeng Z, Deng B, Zhu BS, Cao T, et al. Colorectal cancer (CRC) as a multifactorial disease and its causal correlations with multiple signaling pathways. Biosci Rep. 2020;40(3):BSR20200265. doi: 10.1042/BSR20200265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diao JA, Kohane IS, Manrai AK. Biomedical informatics and machine learning for clinical genomics. Hum Mol Genet. 2018;27(R1):R29–R34. doi: 10.1093/hmg/ddy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barman RK, Mukhopadhyay A, Maulik U, Das S. Identification of infectious disease-associated host genes using machine learning techniques. BMC Bioinform. 2019;20(1):736. doi: 10.1186/s12859-019-3317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen D, Hu J, Zhu M, Tang N, Yang Y, Feng Y. Diagnosis of thyroid nodules for ultrasonographic characteristics indicative of malignancy using random forest. BioData Min. 2020;13:14. doi: 10.1186/s13040-020-00223-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fusar-Poli P, Stringer D, Durieux AMS, Rutigliano G, Bonoldi I, De Micheli A, et al. Clinical-learning versus machine-learning for transdiagnostic prediction of psychosis onset in individuals at-risk. Transl Psychiatry. 2019;9(1):259. doi: 10.1038/s41398-019-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Q, Mao W, Shu Y, Lin F, Liu S, Shen H, et al. A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol. 2012;138(1):85–93. doi: 10.1007/s00432-011-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang ZX, Deng TX, Ma Z. Identification of a 4-miRNA signature as a potential prognostic biomarker for pancreatic adenocarcinoma. J Cell Biochem. 2019;120(10):16416–16426. doi: 10.1002/jcb.28601. [DOI] [PubMed] [Google Scholar]
- 40.Zhong M, Ma X, Sun C, Chen L. MicroRNAs reduce tumor growth and contribute to enhance cytotoxicity induced by gefitinib in non-small cell lung cancer. Chem Biol Interact. 2010;184(3):431–438. doi: 10.1016/j.cbi.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Pei FL, Cao MZ, Li YF. Circ_0000218 plays a carcinogenic role in colorectal cancer progression by regulating miR-139-3p/RAB1A axis. J Biochem. 2020;167(1):55–65. doi: 10.1093/jb/mvz078. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu CW, et al. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer. 2014;13:124. doi: 10.1186/1476-4598-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song M, Yin Y, Zhang J, Zhang B, Bian Z, Quan C, et al. MiR-139-5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell. 2014;5(11):851–861. doi: 10.1007/s13238-014-0093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, et al. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract. 2016;212(7):643–649. doi: 10.1016/j.prp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Xu J, Le VM, Gong Q, Li S, Gao F, et al. EpCAM Aptamer-functionalized cationic liposome-based nanoparticles loaded with miR-139-5p for targeted therapy in colorectal cancer. Mol Pharm. 2019;16(11):4696–4710. doi: 10.1021/acs.molpharmaceut.9b00867. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Ma Y-Y, Wang J, Zeng X-F, Li R, Kang W, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. OncoTargets Ther. 2015;9:139–148. doi: 10.2147/OTT.S95565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan W, Yang W, Liu Z, Wu G. Characterization of microRNA expression in primary human colon adenocarcinoma cells (SW480) and their lymph node metastatic derivatives (SW620) Onco Targets Ther. 2018;11:4701–4709. doi: 10.2147/OTT.S169233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li HT, Zhang H, Chen Y, Liu XF, Qian J. MiR-423-3p enhances cell growth through inhibition of p21Cip1/Waf1 in colorectal cancer. Cell Physiol Biochem. 2015;37(3):1044–1054. doi: 10.1159/000430230. [DOI] [PubMed] [Google Scholar]
- 49.Li X, He X, Wang J, Wang D, Cong P, Zhu A, et al. The regulation of exosome-derived miRNA on heterogeneity of macrophages in atherosclerotic plaques. Front Immunol. 2020;11:2175. doi: 10.3389/fimmu.2020.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, et al. RNA-Seq profiling of serum exosomal circular RNAs reveals Circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol. 2020;10:982. doi: 10.3389/fonc.2020.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho MY, Yu AL, Yu J. Glycosphingolipid dynamics in human embryonic stem cell and cancer: their characterization and biomedical implications. Glycoconj J. 2017;34(6):765–777. doi: 10.1007/s10719-016-9715-x. [DOI] [PubMed] [Google Scholar]
- 52.Iioka H, Saito K, Kondo E. Crumbs3 regulates the expression of glycosphingolipids on the plasma membrane to promote colon cancer cell migration. Biochem Biophys Res Commun. 2019;519(2):287–293. doi: 10.1016/j.bbrc.2019.08.161. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura K, Yamashita K, Sawaki H, Waraya M, Katoh H, Nakayama N, et al. Aberrant methylation of GCNT2 is tightly related to lymph node metastasis of primary CRC. Anticancer Res. 2015;35(3):1411–1421. [PubMed] [Google Scholar]
- 54.Liu JY, Wu XY, Wu GN, Liu FK, Yao XQ. FOXQ1 promotes cancer metastasis by PI3K/AKT signaling regulation in colorectal carcinoma. Am J Transl Res. 2017;9(5):2207–2218. [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Liu Y, Zhang J, Cui X, Li G, Wang J, et al. FOXQ1 promotes gastric cancer metastasis through upregulation of Snail. Oncol Rep. 2016;35(6):3607–3613. doi: 10.3892/or.2016.4736. [DOI] [PubMed] [Google Scholar]
- 56.Zhu Z, Zhu Z, Pang Z, Xing Y, Wan F, Lan D, et al. Short hairpin RNA targeting FOXQ1 inhibits invasion and metastasis via the reversal of epithelial-mesenchymal transition in bladder cancer. Int J Oncol. 2013;42(4):1271–1278. doi: 10.3892/ijo.2013.1807. [DOI] [PubMed] [Google Scholar]
- 57.Pei Y, Wang P, Liu H, He F, Ming L. FOXQ1 promotes esophageal cancer proliferation and metastasis by negatively modulating CDH1. Biomed Pharmacother. 2015;74:89–94. doi: 10.1016/j.biopha.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, et al. FOXQ1 mediates the crosstalk between TGF-β and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther. 2015;16(7):1099–1109. doi: 10.1080/15384047.2015.1047568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor KM, Muraina IA, Brethour D, Schmitt-Ulms G, Nimmanon T, Ziliotto S, et al. Zinc transporter ZIP10 forms a heteromer with ZIP6 which regulates embryonic development and cell migration. Biochem J. 2016;473(16):2531–2544. doi: 10.1042/BCJ20160388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 61.Vecchione A, Baldassarre G, Ishii H, Nicoloso MS, Belletti B, Petrocca F, et al. Fez1/Lzts1 absence impairs Cdk1/Cdc25C interaction during mitosis and predisposes mice to cancer development. Cancer Cell. 2007;11(3):275–289. doi: 10.1016/j.ccr.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou W, He MR, Jiao HL, He LQ, Deng DL, Cai JJ, et al. The tumor-suppressor gene LZTS1 suppresses colorectal cancer proliferation through inhibition of the AKT-mTOR signaling pathway. Cancer Lett. 2015;360(1):68–75. doi: 10.1016/j.canlet.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Sun BL. Current microsatellite instability testing in management of colorectal cancer. Clin Colorectal Cancer. 2020;S1533-0028(20)30104-3. 10.1016/j.clcc.2020.08.001. [DOI] [PubMed]
- 64.de Leon MP, Roncucci L. Microsatellite instability in colorectal cancer. N Engl J Med. 2000;342(21):1607. doi: 10.1056/NEJM200005253422111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. The differential expressed miRNA/mRNA identified by edgeR and limma.
Additional file 2. Table S2. The targeted mRNA of the candidate exosomal miRNAs.
Data Availability Statement
The datasets used and/or analysed during the current study are available from NCBI Gene Expression Omnibus (GEO: GSE39833) https://www.ncbi.nlm.nih.gov/geo/ and the cancer genome database (TCGA: COAD, READ) https://portal.gdc.cancer.gov.