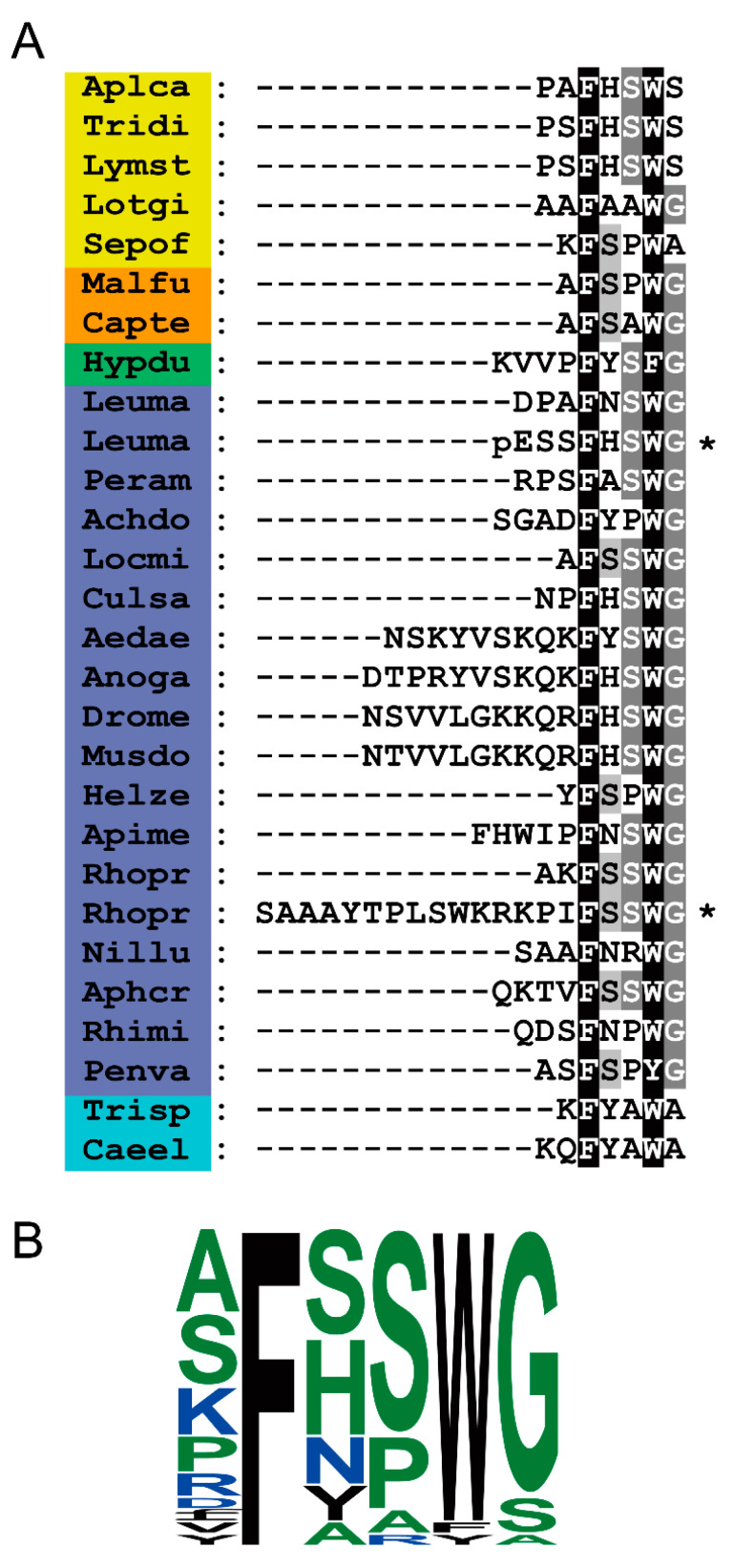
Figure 1.
Sequences of leucokinins. (A) Sequence alignments of leucokinin and leucokinin-like peptides from select species. Note that C-terminal amidation is not shown. Conserved residues are highlighted in black (identical) or gray (similar). Species belonging to the same phyla have been highlighted with the same color. Species names are as follows, Mollusks: Aplca (Aplysia californica), Tridi (Tritonia diomedea), Lymst (Lymnaea stagnalis), Lotgi (Lottia gigantea), Sepof (Sepia officinalis). Annelids: Malfu (Malacoceros fuliginosus), Capte (Capitella teleta). Tardigrade: Hypdu (Hypsibius dujardini). Insects: Leuma (Leucophaera maderae), Peram (Periplaneta americana), Achdo (Acheta domesticus), Locmi (Locusta migratoria), Culsa (Culex salinarius), Aedae (Aedes aegypti), Anoga (Anopheles gambiae), Drome (Drosophila melanogaster), Musdo (Musca domestica), Helze (Helicoverpa zea), Apime (Apis mellifera), Rhopr (Rhodnius prolixus), Nillu (Nilaparvata lugens), Aphcr (Aphis craccivora). Tick (Ixodida): Rhimi (Rhipicephalus microplus). Shrimp (Crustacea): Penva (Penaeus vannamei). Nematodes: Trisp (Trichinella spiralis), and Caeel (Caenorhabditis elegans). Note that the T. spiralis and C. elegans peptides are unlikely to be LKs as no canonical LK precursor was found (see also text). Sequences indicated by asterisks (*) have special features—in L. maderae it is N-terminally pyroglutamate blocked (pE) and in R. prolixus, the peptide is N-terminally extended. (B) Sequence logo of the LK peptides in different species.