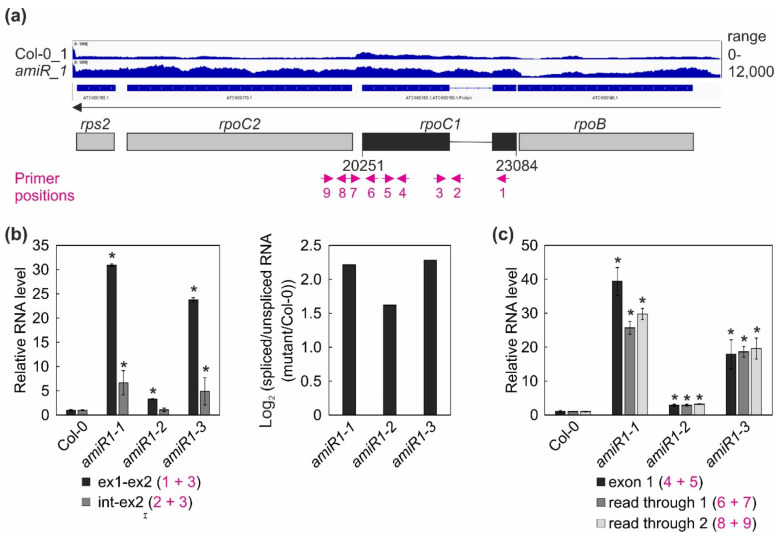
Figure 7.
Expression and processing of chloroplast rpoC1 transcripts in wild-type (Col-0) and mterf2 mutant plants. (a) RNA-Seq analysis was performed with RNA extracted from wild-type (Col-0) and amiR1-3-mterf2 mutant (amiR) seedlings. The read depths of rps2, rpoC2, rpoC1, and rpoB transcripts detected in Col-0 and amiR seedlings were visualized with the Integrative Genomics Viewer (IGV). Below the IGV data, a schematic representation of the chloroplast rpoC1 gene is shown and the magenta arrows indicate the primer positions used in Figure (b). (b,c) Quantitative RT-PCR analysis of RNAs isolated from 6-day-old Col-0 and amiR1–mterf2 mutant plants (amiR1-1, amiR1-2, and amiR1-3). PCR was conducted with the primer pairs marked in panel (a) and AT4G36800, encoding an RUB1–conjugating enzyme (RCE1) as a control. The RNA expression levels are reported relative to that in the Col-0, which was set to 1. The ratios of spliced to unspliced rpoC1 transcript between amiR1–mterf2 mutant seedlings (amiR1-1, amiR1-2, and amiR1-3) and wild-type (Col-0) were calculated based on the relative levels of rpoC1 exon 1, exon 2, and the intervening intron, and the data are depicted as the log2 ratio of spliced to unspliced rpoC1 transcript in the amiR1–mterf2 mutant plants compared to Col-0 in (b). Relative RNA levels of exon 1, read through 1 and -2 in Col-0 and amiR1–mterf2 mutant plants are shown in (c). Bars indicate standard deviations (SD). Significant differences between mutants and Col-0 were evaluated with Student’s t-test (P < 0.05) and they are denoted by the asterisks (*).