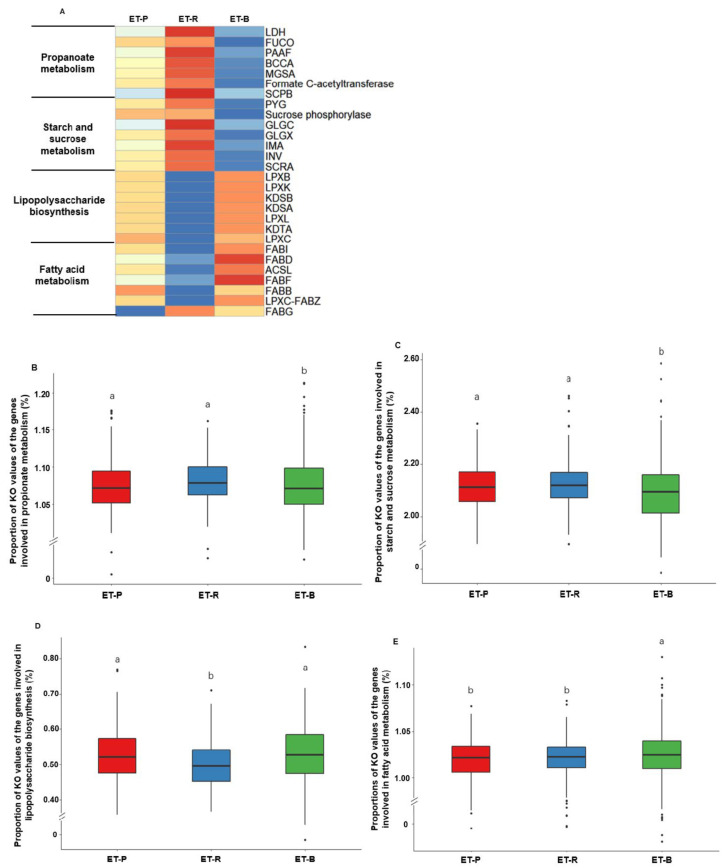
Figure 6.
Heatmaps of abundant genes involved in metabolic functions determined by (A) PICRUSt2 and (B–E) the relative abundance of the genes related to significant metabolic functions in three enterotypes. (A) The association of the genes related to different functions in three enterotypes. (B) Propionate metabolism in three enterotypes. (C) Starch and sucrose metabolism in three enterotypes. (D) Lipopolysaccharide biosynthesis in three enterotypes. (E) Fatty acid metabolism in three enterotypes. LDH, L-lactate dehydrogenase; Figure 1. phosphate adenylyltransferase; GLGX, glycogen debranching enzyme; IMA, oligo-1,6-glucosidase; INV, beta-fructofuranosidase; SCRA, sucrose phosphoenolpyruvate-dependent sucrose phosphotransferase system permease component; LPXB, lipid-A-disaccharide synthase; LPXK, tetraacyldisaccharide 4′-kinase; KDSB, 3-deoxy-mannooctulosonate cytidylyltransferase; KDSA, 2-dehydro-3-deoxyphosphooctonate aldolase; LPXL, Kdo2-lipid IVA lauroyltransferase/acyltransferase; KDTA, 3-deoxy-D-manno-octulosonic-acid transferase; LPXC, UDP-3-O-(3-hydroxymyristoyl) N-acetylglucosamine deacetylase; FABI, enoyl-[acyl-carrier protein] reductase I; FABD, [acyl-carrier-protein] S-malonyltransferase; ACSL, long-chain acyl-CoA synthetase; FABF, 3-oxoacyl-[acyl-carrier-protein] synthase II; FABB, 3-oxoacyl-[acyl-carrier-protein] synthase I; LPXC-FABZ_2, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase/3-hydroxyacyl-[acyl-carrier-protein] dehydratase; FABG, 3-oxoacyl-[acyl-carrier protein] reductase. KO, KEGG Orthologues. Different alphabets (a,b,c) represented significant differences among the different bacteria in the family level by Tukey’s test at p < 0.05. ET-P, Prevotellaceae; ET-R, Ruminococcaceae; ET-B, Bacteroidaceae.