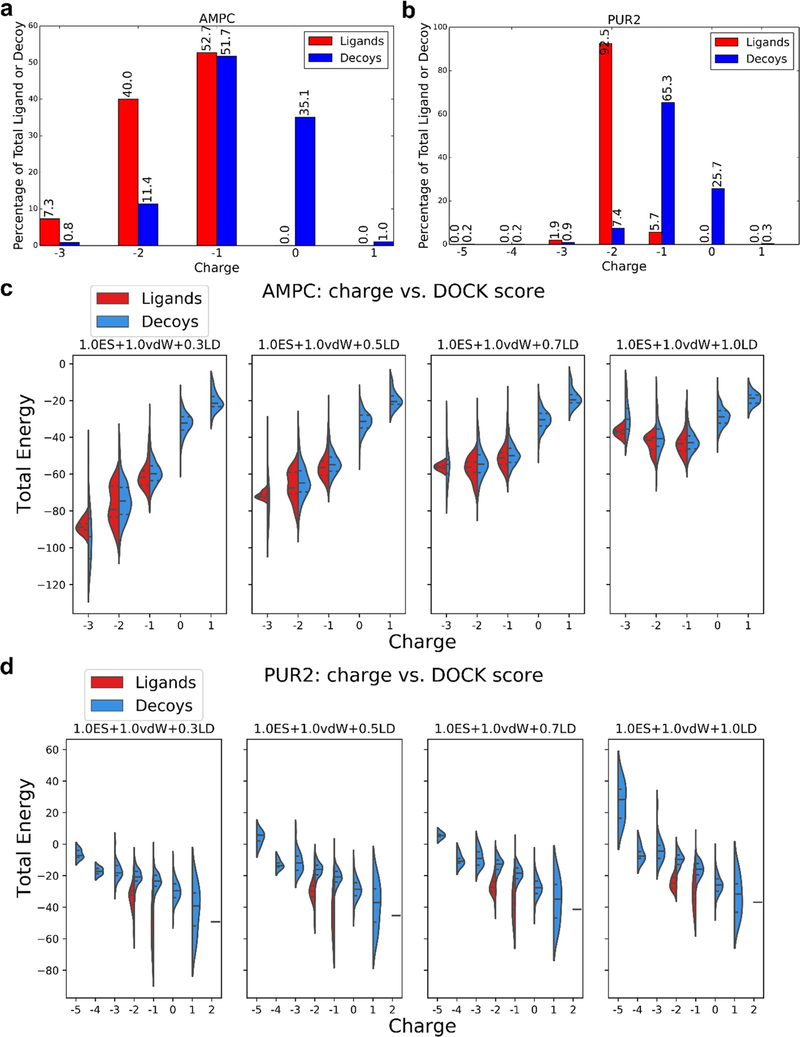
Figure 2.
Proportion of charged ligands and decoys in the DUD-E benchmarking sets coupled with altered electrostatic and ligand desolvation weights affects the DOCK energies and thus LogAUC values. Percentage of ligands or decoys in the DUD-E set with a given charge for AmpC β-lactamase (AmpC, a) and GAR transformylase (PUR2, b). Comparison of DOCK energy and molecule charge for AmpC β-lactamase (AmpC, c) and GAR transformylase (PUR2, d) for the electrostatic coefficient of 1.0 and the four ligand desolvation weights (0.3, 0.5, 0.7, and 1.0). Central dotted lines of DOCK energies represent the medians, upper dotted lines represent the third quartiles, and lower dotted lines represent the first quartiles for both scoring functions. The lowest points represent the minimum DOCK energies, and the highest values represent the maximum DOCK energies. The AmpC ligands in DUD-E are predominantly anionic (a), and while this is also true for the decoys, the latter harbors a higher ratio of neutral molecules. Increasing the ligand desolvation coefficient ranks neutral molecules higher (as sorted by total DOCK energy), favoring decoys, and enrichment decreases (c). Conversely, increasing the electrostatic coefficient favors the anionic ligands, increasing the enrichment. The large majority of PUR2 ligands is di-anionic, while the decoys are monoanionic (b), providing an advantage to the ligands at lower ligand desolvation coefficients (as sorted by total DOCK energy) (d), as they can form more favorable electrostatic interactions with the protein without a large ligand desolvation cost.