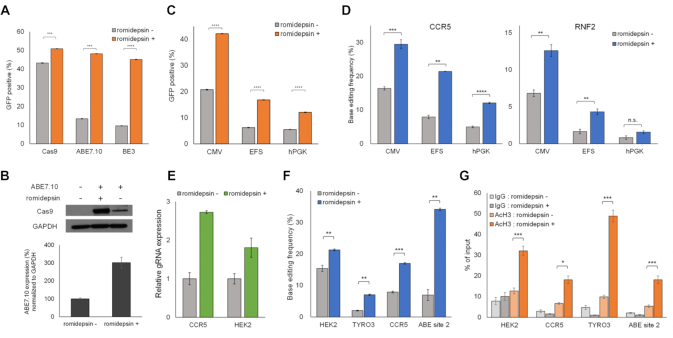
Figure 3.
Romidepsin improves base editing efficiency by affecting the protein and gRNA expression levels and the chromatin state. (A) Flow cytometry analysis of the expression levels of ABE7.10–2A-EGFP, spCas9–2A-EGFP and BE3–2A-EGFP proteins in the presence and absence of romidepsin. The proportion of GFP-positive cells increased following romidepsin treatment. (B) Western blot assay to demonstrate the ABE7.10 protein expression levels in HEK293T/17 cells. The expression levels of ABE7.10 proteins increased in the presence of romidepsin. GAPDH is shown as a loading control. (C) Flow cytometry analysis of the expression levels of CMV, EFS, and hPGK promoter-driven ABE7.10-2A-EGFP proteins in the presence and absence of romidepsin. Romidepsin improved both EFS and hPGK promoter-driven protein expression. (D) Targeted deep sequencing analysis showed that romidepsin improved the base editing efficiency at both CCR5 and RNF2 sites in HEK293T/17 cells transfected with EFS- and hPGK promoter-driven ABE7.10 expressing plasmids. (E) Quantitative real-time PCR analysis for detecting gRNA expression levels. Romidepsin increased U6 promoter-driven gRNA expression levels. (F) Base editing efficiencies at four endogenous target sites in RNP delivered HEK293T/17 cells. (G) ChIP assay results at four endogenous target sites. The acetylation percentage at the four sites increased with romidepsin treatment. Error bars indicate SEM (n = 3); ns, not significant; P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (using two-tailed Student's t-test).