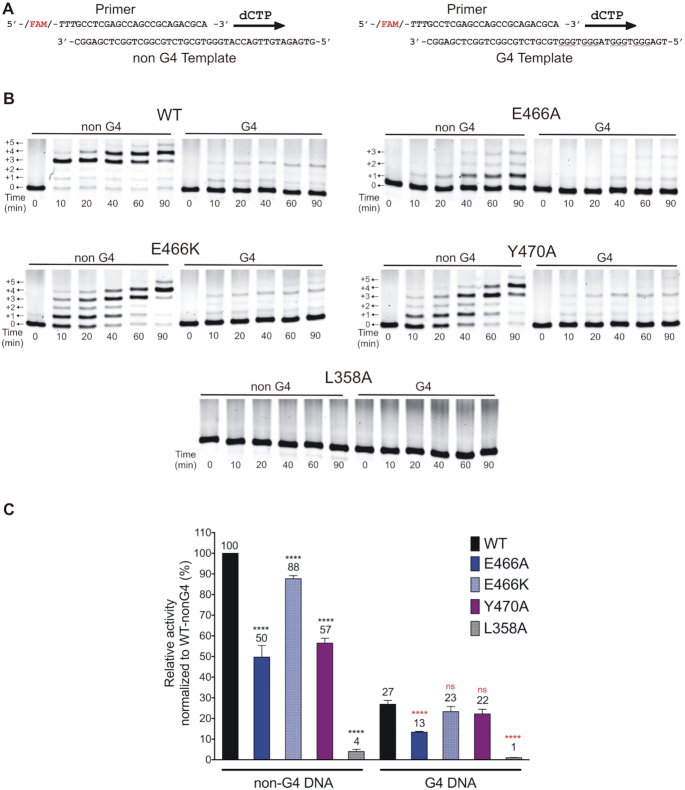
Figure 4.
Alterations in the catalytic activity of hRev1 mutants were largely unrelated to whether G4 was present in the template strand. (A) Schematic illustration of the primer-template DNA substrates. Arrows indicate the direction of primer extension by multiple insertions of dCMP across the template during the time course. (B) Representative gel images for the time course monitoring dCMP insertion by the wild-type and mutant hRev1330–833 proteins (50 nM) on non-G4 or G4 DNA substrates (200 nM) in a buffer containing 100 mM KCl. The positions of the primer and additions of dCMP (+1, +2, etc.) are marked beside the gel images. (C) Relative enzyme activity for each protein is shown for the non-G4 and G4 DNA substrates. Relative activity is reported as a percentage of the activity of wild-type hRev1330–833 on the non-G4 DNA substrate. The mean value for relative activity for each protein is shown at the top of its corresponding bar. Reported values represent the mean ± SD (n = 3). Statistical significance was assessed by performing a one-way ANOVA with multiple comparisons, and the P-values were calculated for each comparison. **** indicates P < 0.0001 for comparisons between mean values of each mutant enzyme with that of the wild-type on non-G4 substrate, **** indicates P < 0.0001 for comparisons between mean values of each mutant enzyme with that of the wild-type on the G4 substrate. ns; non-significant (P> 0.05).