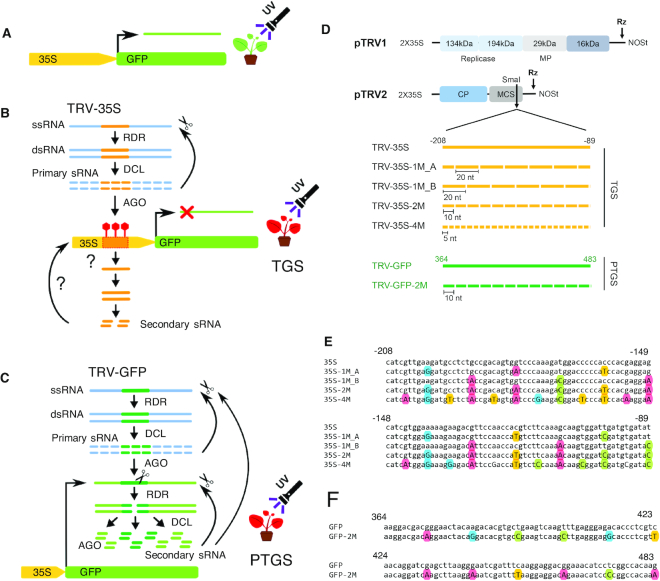
Figure 1.
TRV-based VIGS system used for studying the impact of mismatched small RNAs on silencing of the GFP transgene. (A) Schematic diagram of the CaMV 35S promoter driven GFP transgene in N. benthamiana 16c plant. 16c plants show green fluorescence under UV light. (B) Schematic diagram of virus induced TGS. Reproduction of RNA viruses, such as TRV results in the accumulation of double-stranded replication intermediates, which are processed into primary sRNAs by antiviral DICER-like (DCL) nucleases. sRNAs are then associated with and guide ARGONAUTE (AGO) proteins to nucleic acid targets by base-pair complementarity. If the target is chromatin, sRNAs can induce the methylation of cytosine residues. This process is referred to as RdDM. Consequently, inoculation of 16c plants with a recombinant TRV carrying the 35S promoter sequence (TRV-35S) can bring about RdDM, which results in TGS of the GFP reporter gene. GFP silenced 16c plants display red fluorescence under UV-light due the autofluorescence of chlorophyl in lack of GFP expression. It is not known whether RdDM of 35S is associated with the production of secondary (target-generated) sRNAs. sRNA-induced DNA methylation is indicated as red lollipop. RDR, RNA-dependent RNA polymerase (C) Schematic diagram of virus induced PTGS. Virus-derived primary sRNAs can also be loaded into AGO complexes to destroy target RNAs with complementary sequences. It is known as antiviral PTGS, which involves AGO-induced cleavage, destabilization, or translational inhibition. Infecting 16c plants with a recombinant TRV harbouring the GFP coding sequence (TRV-GFP) can result in PTGS of both TRV and GFP mRNAs. Intriguingly this process is associated with the generation of secondary (GFP-specific) sRNA. sRNA-induced cleavage is indicated as scissors. (D) Schematic diagram of the TRV VIGS vectors pTRV1 and pTRV2. A 120 nt fragment of the CaMV 35S promoter (-208 to -89 relatives to the transcription start site, yellow lines) or a 120 nt fragment of the GFP coding sequence (+364 to +483, green lines) was cloned into pTRV2 to induce TGS and PTGS, respectively. Single nucleotide substitutions (SNS; white boxes) were introduced into the 120 nt fragments at regular intervals of 20, 10 or 5 nucleotides, which produced sRNAs with one, two or four mismatches, respectively. SNS were introduced from position 10 in TRV-35S-1M_A and from position 20 in TRV-35S-1M_B. pTRV1 was used along with pTRV2 to generate functional TRV particles. Rz, self-cleaving ribozyme; MCS, multiple cloning sites; CP, coat protein; MP, movement protein; NOSt, NOS terminator. (E) Sequence alignment of the 120 nt fragment from CaMV 35S and its derivatives from (A). Substituted A, C, G, T nucleotides are highlighted with red, green, blue and yellow coloured circles, respectively. (F) Sequence alignment of the 120 nt fragment from GFP5 and its derivative from (A).