Figure 8.
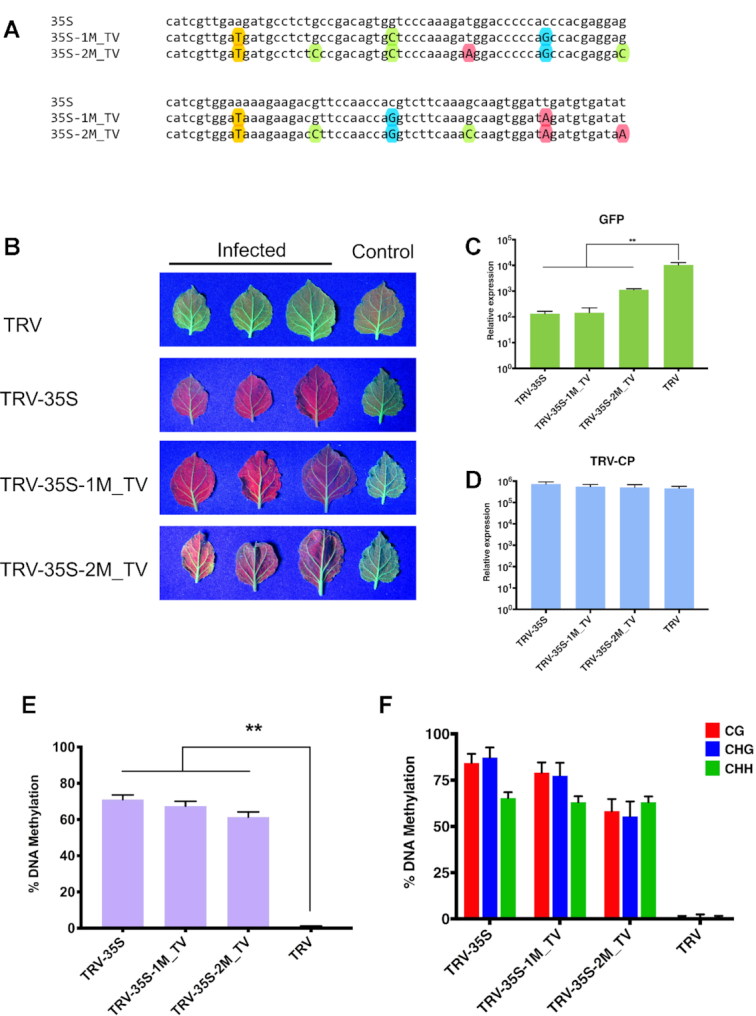
Small RNAs with transversal mismatches can also induce strong TGS. (A) Sequence alignment of the 120 nt fragment from CaMV 35S (from -208 to -89) and its derivatives. Substituted A, C, G, T nucleotides are highlighted with red, green, blue and yellow coloured circles, respectively. (B) Systemic leaves of N. benthamiana 16c plants infected with recombinant TRV as indicated. Leaves were collected from independent plants. An uninfected 16c leaf is shown as a control (right). Leaves were photographed at 21 dpi under UV light. (C and D) Analysis of GFP expression and TRV accumulation in recombinant TRV-infected plants by qRT-PCR. RNA was extracted from the systemically infected leaves at 21 dpi. Error bars show the standard error of the mean (SEM) of three independent biological replicates. Asterisks indicate significant differences (Student's t-test, P < 0.01). (E) Analysis of DNA methylation at the target CaMV 35S promoter (from -208 to -89) by bisulfite sequencing in TRV-infected N. benthamiana 16c plants. DNA was extracted from the systemically infected leaves at 21 dpi. The histogram shows the percentage of total methylated cytosine. Asterisks indicate significant differences (Student's t-test, P < 0.01). (F) Summary of bisulfite sequencing analysis. Red, blue and green bars indicate the percentage of methylated cytosine residues at CG, CHG and CHH sites, repetitively. Results presented in (E) and (F) were obtained from three independent biological replicates. Raw data are available in Supplementary Figure S10. Error bars represent a 95% interval (Wilson score interval; see details in Supplementary Figure S10B).
