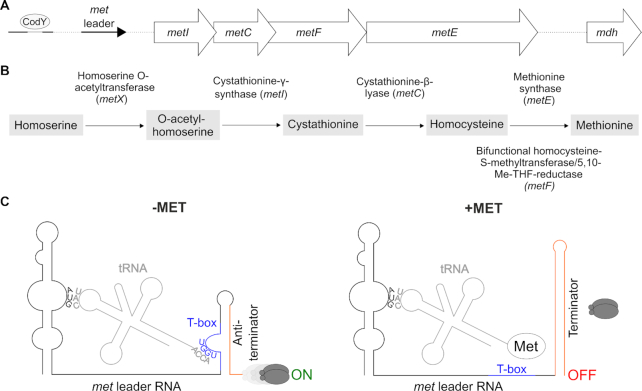
Figure 1.
(A) Schematic view of the organization of the S. aureus met operon including a CodY binding site and its 5′ UTR (met leader). (B) Methionine biosynthesis pathway in S. aureus, Me-THF, methylene-tetrahydrofolate. (C) Schematic of the binding interactions between tRNA and met leader RNA (T-box riboswitch). Left: system under methionine deprivation (‘−MET’), right: system under high intracellular methionine levels (‘+MET’). Base pairing of tRNA anticodon (‘CAU’) with specifier codon (‘AUG’) of met leader and of free 3′ tRNA end (‘ACCA’) with T-box (depicted in blue) sequence (‘UGGU’) is shown. Terminator sequence is highlighted in orange, gray ellipses symbolize RNA polymerase, ‘ON’: read-through into downstream genes, met mRNA transcription, ‘OFF’: premature transcription termination, no met mRNA transcription.