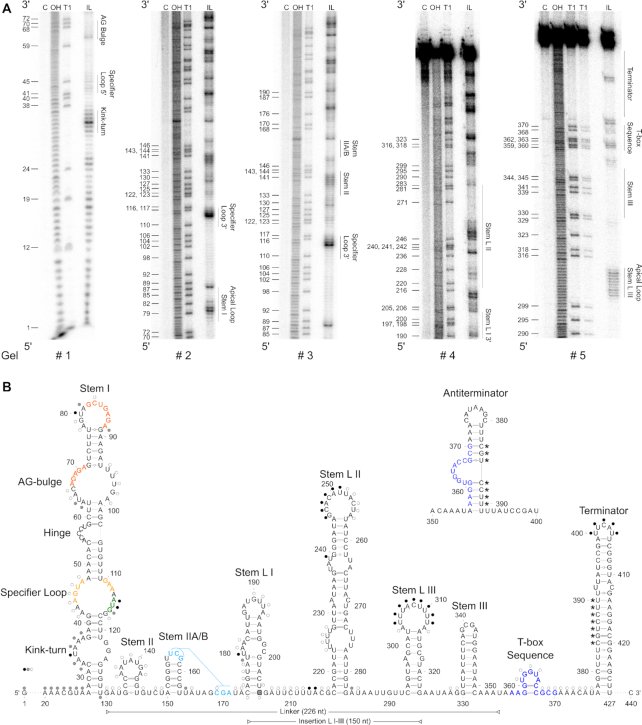
Figure 2.
Secondary structure of met leader. (A) In-line probing PAA gels used to build 2D model. Nomenclature of structural motifs according to (9). Additional stems within linker region are numbered L I to L III. Position of guanosines (G) is given on the left of each gel. Structural motifs are specified on right side of each gel. C: control reaction; untreated RNA, OH: alkaline hydrolysis reaction; ladder, T1: RNase T1-treated RNA; G-specific ladder, IL: in-line reaction. (B) 2D structure model of met leader in its OFF state, predicted antiterminator 2D structure is shown above. Color of dots (black, gray, white) next to nucleotides indicates cleavage intensity (high, medium, low) detected in (A). Interacting nucleotides of specifier loop are shown in yellow, base pairing nucleotides of AG-bulge and apical loop of stem I in orange, specifier codon in green, potentially interacting nucleotides of stem IIA/B pseudoknot in light blue, T-box sequence in dark blue. Linker region between stem I and T-box sequence and insertion region of stem L I–III are indicated by horizontal bars. Nucleotides of terminator base pairing with T-box sequence in antiterminator conformation are marked by asterisks (*). G 205 highlighted in gray indicates 5′ end of short met leader RNA used for in-line probing.