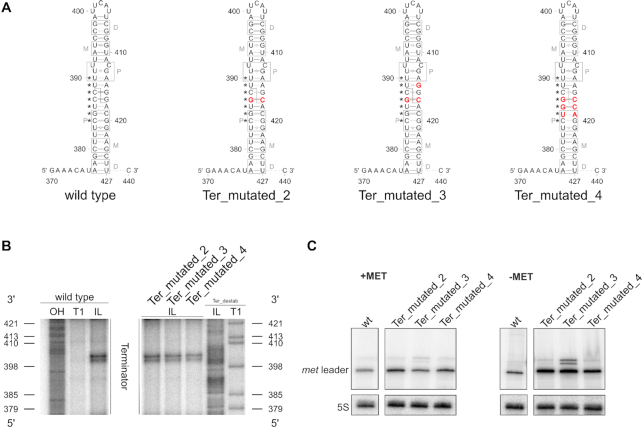
Figure 6.
Sequence constraints of met leader RNA terminator cleavage. (A) Sequence and predicted secondary structure of terminator regions in met leader RNAs of wild type and mutants ‘Ter_mutated_2’, ‘Ter_mutated_3’ and ‘Ter_mutated_4’. Point mutations introduced are highlighted in red, gray boxes indicate regions interacting with RNase III dimers as described in (40), P: proximal, M: middle, D: distal box. Nucleotides engaged to form the T-box bulge in antiterminator conformation (see also Figure 2B) are marked by asterisks (*). Position of RNase III cleavage site in wild type met leader is indicated by a dashed line. (B) In-line probing gel sections of the terminator region of met leader wild type, ‘Ter_mutated_2’, ‘Ter_mutated_3’ and ‘Ter_mutated_4’ RNA are shown. Position of guanosines (G) as in ‘Ter_destab’ is given on the left and right of the gel, respectively, annotations as in Figure 2A. (C) Total RNA isolated from S. aureus Newman and the met leader mutants ‘Ter_mutated_2’, ‘Ter_mutated_3’ and ‘Ter_mutated_4’ grown in CDM with (‘+MET’) or without methionine (‘−MET’) was run on a PAA gel. Northern blot was hybridized with a met leader-specific probe and subsequently re-hybridized with a 5S rRNA-specific probe as loading control.