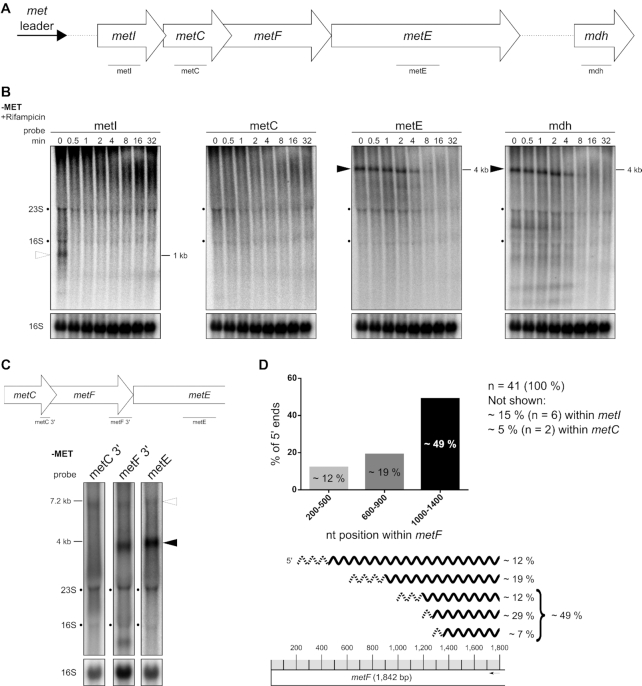
Figure 7.
Stability of the met operon mRNA varies over length of the transcript. (A) Schematic view of the organization of the met operon including its 5′ UTR (met leader). Lines below gene arrows indicate relative positions of the respective probes used in (B). (B) Total RNA isolated from S. aureus Newman grown in CDM without methionine (‘−MET’) over a time course after rifampicin addition (0–32 min) was run on an agarose gel. Northern blot was probed for metI, metC, metE and mdh. Open arrowhead marks metI mRNA and black arrowheads mark 3′ met mRNA. Approximate transcript lengths are indicated on the right of the respective blot. Re-hybridization with a 16S rRNA-specific probe was used as loading control. (C) Schematic view of the met operon 3′ region (without mdh) and relative positions of probes used. Total RNA isolated from S. aureus Newman grown in CDM without methionine (‘−MET’) was run on an agarose gel. Northern blot was probed for 3′ region of metC (‘metC 3′’), 3′ region of metF (‘metF 3′’) and metE. Open arrowhead marks full-length met operon mRNA (without met leader) and black arrowhead marks 3′ met operon mRNA. Approximate transcript lengths are indicated on the left. Re-hybridization with a 16S rRNA-specific probe was used as loading control. (D) Summary of 5′-RACE data. Upper diagram: Percentage of 5′ ends detected within distinct regions of metF. Lower part: Mapping of the detected 5′ ends to the metF region. Scale of the metF gene is illustrated at the bottom. Gray boxes represent 100 nucleotides each and the black arrow marks the approximate position of the primer used for cDNA synthesis. Transcripts characterized by 5′-RACE are depicted as wavy lines, dashed regions symbolize the range of detected 5′ ends. The percentage of each transcript group detected is given on the right.