Figure 8.
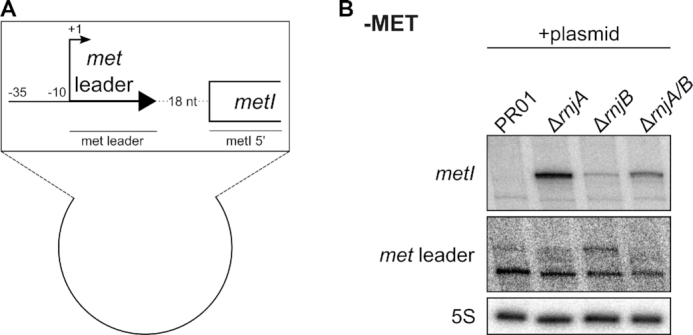
RNase J is involved in met operon mRNA degradation. (A) Scheme of the plasmid transformed into S. aureus SA564RD ΔpyrFE (‘PR01’) and its isogenic RNase J1 (‘ΔrnjA’), RNase J2 (‘ΔrnjB’) and RNase J1/J2 double (‘ΔrnjA/B’) mutants. ‘–35’ and ‘–10’ indicates the promoter region. Arrow with +1 marks the transcription start site of met leader. The met leader sequence is depicted as thick, black arrow. First 215 nt of metI are shown as open rectangle. Lines below genes indicate relative positions of probes used in (B). (B) Total RNA was isolated from S. aureus SA564RD ΔpyrFE (‘PR01’) and its isogenic RNase J1 (‘ΔrnjA’), RNase J2 (‘ΔrnjB’) and RNase J1/J2 double (‘ΔrnjA/B’) mutants. Bacteria were grown in MH medium to OD 0.5. Then cultures were washed twice with PBS, bacteria were shifted into CDM without methionine (‘−MET’) supplemented with pyrimidine for 30 min and samples were taken. 5 μg of total RNA was run on a PAA gel. Northern blot was hybridized with a met leader-specific probe, re-probed with a metI 5′-specific probe and subsequently re-hybridized with a 5S rRNA-specific probe as loading control.
