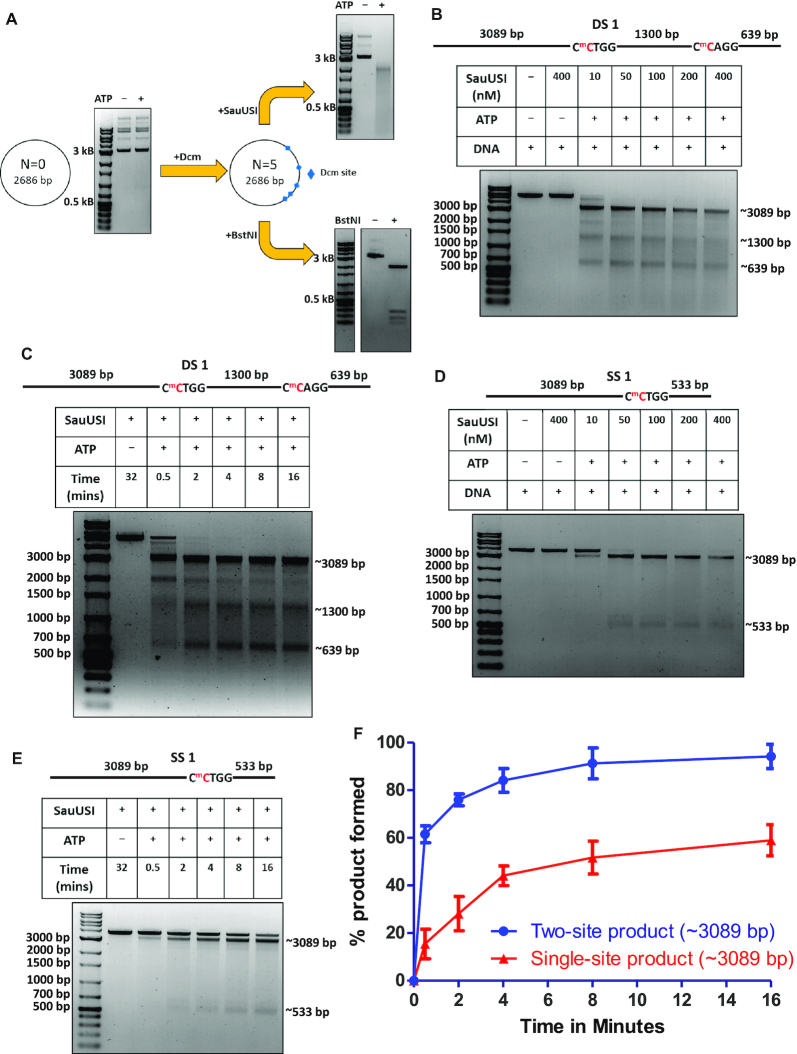
Figure 1.
Nuclease activity of SauUSI. (A) Schematic of the plasmid used for cleavage. N represents the number of target sites of SauUSI in a particular plasmid. Methylation was brought about in vivo to generate the target sites of SauUSI. The corresponding cleavage pattern (on a 1% agarose gel) of SauUSI on the respective plasmids. The reactions with and without nucleotide (ATP) are compared against a DNA marker. (B) The two-site substrate (DS 1) used for the cleavage assay; the separation between the two sites is 1300 bp. The cytosine highlighted in red represents methylation. The target site is methylated on both the strands. Representative 1% agarose gel for two-site cleavage using varying concentrations (10–400 nM) of SauUSI. (C) Single-site substrate (SS 1) used for the cleavage assay. The cytosine highlighted in red represents methylation. Methylation is present on both the strands. The target site is methylated on both the strands. Representative 1% agarose gel for single-site cleavage using varying concentrations (10–400 nM) of SauUSI. (D) Representative 1% agarose gel for two-site cleavage (DS 1) carried over a time period of 30 s to 16 min. (E) Representative 1% agarose gel for single-site (SS 1) cleavage carried over a time period of 30 seconds to 16 min. (F) Percentage of product (the ∼3089 bp band) formed as a function of time (DNA concentration 3 nM and protein concentration 100 nM) (n = 3). In the case of DS 1, the ∼3089 bp fragment can arise from a single-site cleavage (at the left hand side) or from a sequential two-site cleavage.