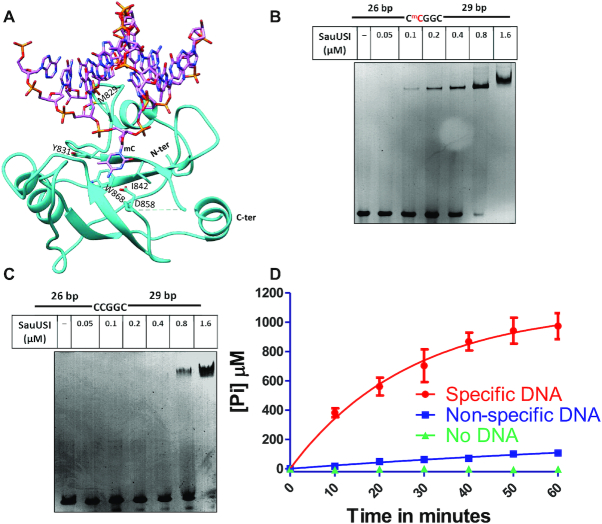
Figure 4.
Target recognition by the SRA domain and DNA dependent ATP stimulation of SauUSI. (A) Structure of the SRA domain of SauUSI with DNA modeled. The DNA is from the structure of the structural homolog SUVH6, an H3K9 histone methyltransferase from Arabidopsis thaliana (PDB ID: 6A5N). (B) The specific DNA substrate used for EMSA. The cytosine highlighted in red represents methylation. The target site is methylated on both the strands. Representative 5% native PAGE EMSA gel using specific DNA substrate (250 nM) and assay performed over a protein concentration range of 0.05–1.6 μM. (C) The non-specific DNA substrate used for EMSA. Representative 5% native PAGE EMSA gel using non-specific DNA substrate (250 nM) and assay performed over a protein concentration range of 0.05–1.6 μM. (D) ATPase stimulation assay carried out using the malachite green method, represented by inorganic phosphate released (on the Y-axis) over time (on the X-axis) in reactions containing specific DNA, non-specific DNA and no DNA (n = 3).